No Slide Title
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Inside: Idaea Asceta (Prout) (Geometridae), New to the U.S
________________________________________________________________________________________ Volume 53, Number 3 Fall 2011 www.lepsoc.org ________________________________________________________________________________________ Inside: Idaea asceta (Prout) (Geometridae), new to the U.S. Lepidoptera conserva- tion under a changing climate Karl Jordan Award win- ner: Don Lafontaine Life history of Leona’s Little Blue Tiputini Biological Sta- tion, Ecuador Late Season trip to the Richardson Mountains Membership Updates, The Mailbag, Marketplace... ... and more! ________________________________________________________________________________________ ________________________________________________________ Contents ________________________________________________________www.lepsoc.org A Late Season Trip to the Richardson Mountains ____________________________________ Michael Leski. ............................................................................................ 75 Volume 53, Number 3 Report on the Southern Lepidoptersists’ Society and Association Fall 2011 for Tropical Lepidoptera Meeting 2011 The Lepidopterists’ Society is a non-profit ed- Jacqueline Y. Miller. ............................................................................. 78 ucational and scientific organization. The ob- Idaea asceta (Prout) (Geometridae: Sterrhinae) from Texas, new ject of the Society, which was formed in May to the North American fauna 1947 and formally constituted in December Charles V. Covell. ................................................................................... -

Reading the Complex Skipper Butterfly Fauna of One Tropical Place
Reading the Complex Skipper Butterfly Fauna of One Tropical Place Daniel H. Janzen1*, Winnie Hallwachs1, John M. Burns2, Mehrdad Hajibabaei3, Claudia Bertrand3, Paul D. N. Hebert3 1 Department of Biology, University of Pennsylvania, Philadelphia, Pennsylvania, United States of America, 2 Department of Entomology, National Museum of Natural History, Smithsonian Institution, Washington, D.C., United States of America, 3 Department of Integrative Biology, Biodiversity Institute of Ontario, University of Guelph, Guelph, Canada Abstract Background: An intense, 30-year, ongoing biodiversity inventory of Lepidoptera, together with their food plants and parasitoids, is centered on the rearing of wild-caught caterpillars in the 120,000 terrestrial hectares of dry, rain, and cloud forest of Area de Conservacion Guanacaste (ACG) in northwestern Costa Rica. Since 2003, DNA barcoding of all species has aided their identification and discovery. We summarize the process and results for a large set of the species of two speciose subfamilies of ACG skipper butterflies (Hesperiidae) and emphasize the effectiveness of barcoding these species (which are often difficult and time-consuming to identify). Methodology/Principal Findings: Adults are DNA barcoded by the Biodiversity Institute of Ontario, Guelph, Canada; and they are identified by correlating the resulting COI barcode information with more traditional information such as food plant, facies, genitalia, microlocation within ACG, caterpillar traits, etc. This process has found about 303 morphologically defined species of eudamine and pyrgine Hesperiidae breeding in ACG (about 25% of the ACG butterfly fauna) and another 44 units indicated by distinct barcodes (n = 9,094), which may be additional species and therefore may represent as much as a 13% increase. -

A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico. Gary Noel Ross Louisiana State University and Agricultural & Mechanical College
Louisiana State University LSU Digital Commons LSU Historical Dissertations and Theses Graduate School 1967 A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico. Gary Noel Ross Louisiana State University and Agricultural & Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_disstheses Recommended Citation Ross, Gary Noel, "A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico." (1967). LSU Historical Dissertations and Theses. 1315. https://digitalcommons.lsu.edu/gradschool_disstheses/1315 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Historical Dissertations and Theses by an authorized administrator of LSU Digital Commons. For more information, please contact [email protected]. This dissertation has been microfilmed exactly as received 67-14,010 ROSS, Gary Noel, 1940- A DISTRIBUTIONAL STUDY OF THE BUTTERFLIES OF THE SIERRA DE TUXTLA IN VERACRUZ, MEXICO. Louisiana State University and Agricultural and Mechanical CoUege, Ph.D., 1967 Entomology University Microfilms, Inc., Ann Arbor, Michigan A DISTRIBUTIONAL STUDY OF THE BUTTERFLIES OF THE SIERRA DE TUXTLA IN VERACRUZ, MEXICO A D issertation Submitted to the Graduate Faculty of the Louisiana State University and A gricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Department of Entomology by Gary Noel Ross M.S., Louisiana State University, 196*+ May, 1967 FRONTISPIECE Section of the south wall of the crater of Volcan Santa Marta. May 1965, 5,100 feet. ACKNOWLEDGMENTS Many persons have contributed to and assisted me in the prep aration of this dissertation and I wish to express my sincerest ap preciation to them all. -

Saturniidae from Santa Catarina State, Brazil, with Taxonomic Notes (Lepidoptera) 215-220 Nachr
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Nachrichten des Entomologischen Vereins Apollo Jahr/Year: 2009 Band/Volume: 30 Autor(en)/Author(s): Siewert Ricardo R., Silva Eduardo J. E., Mielke Carlos G. C. Artikel/Article: Saturniidae from Santa Catarina State, Brazil, with taxonomic notes (Lepidoptera) 215-220 Nachr. entomol. Ver. Apollo, N. F. 30 (4): 215–220 (2010) 215 Saturniidae from Santa Catarina State, Brazil, with taxonomic notes (Lepidoptera) Ricardo R. Siewert, Eduardo J. E. Silva and Carlos G. C. Mielke Ricardo Russo Siewert & Eduardo José Ely e Silva, Departamento de Fitossanidade, Faculdade de Agronomia Eliseu Maciel, Universidade Federal de Pelotas (UFPEL), Campus Universitário, s/nº, caixa postal 354, CEP, 96010900, Pelotas, RS, Brasil; [email protected] & [email protected] Carlos Guilherme Costa Mielke, Caixa Postal 1206, 84.145000 Carambeí, Paraná, Brasil; [email protected] Abstract: A species list of the Saturniidae (Lepidoptera) of Material and methods the state of Santa Catarina, Brasil, is presented. 149 species are listed in four subfamilies: Arsenurinae (16), Ceratocam Collections visited are listed below (all in Brazil) with pinae (32), Hemileucinae (90), and Saturniinae (11). The their abbreviations, besides their code used in Table 1: following are stat. rev. as species: Hylesia corevia (Hüb ner, CGCM (= X1) Col. Carlos G. C. Mielke, Curitiba, Paraná. [1825]) and Eacles lauroi Oiticica, 1938. CMN (= X2) Museu Nacional, Rio de Janeiro, Rio de Janeiro. Key words: fauna survey, taxonomy, neotropical. DZUP (= X3) Col. Padre Jesus S. Moure, Departamento de Zoo Saturniidae aus Santa Catarina, Brasilien mit logia, Universidade Federal do Paraná, Curitiba, Pa ra ná. -

INSECTA: LEPIDOPTERA) DE GUATEMALA CON UNA RESEÑA HISTÓRICA Towards a Synthesis of the Papilionoidea (Insecta: Lepidoptera) from Guatemala with a Historical Sketch
ZOOLOGÍA-TAXONOMÍA www.unal.edu.co/icn/publicaciones/caldasia.htm Caldasia 31(2):407-440. 2009 HACIA UNA SÍNTESIS DE LOS PAPILIONOIDEA (INSECTA: LEPIDOPTERA) DE GUATEMALA CON UNA RESEÑA HISTÓRICA Towards a synthesis of the Papilionoidea (Insecta: Lepidoptera) from Guatemala with a historical sketch JOSÉ LUIS SALINAS-GUTIÉRREZ El Colegio de la Frontera Sur (ECOSUR). Unidad Chetumal. Av. Centenario km. 5.5, A. P. 424, C. P. 77900. Chetumal, Quintana Roo, México, México. [email protected] CLAUDIO MÉNDEZ Escuela de Biología, Universidad de San Carlos, Ciudad Universitaria, Campus Central USAC, Zona 12. Guatemala, Guatemala. [email protected] MERCEDES BARRIOS Centro de Estudios Conservacionistas (CECON), Universidad de San Carlos, Avenida La Reforma 0-53, Zona 10, Guatemala, Guatemala. [email protected] CARMEN POZO El Colegio de la Frontera Sur (ECOSUR). Unidad Chetumal. Av. Centenario km. 5.5, A. P. 424, C. P. 77900. Chetumal, Quintana Roo, México, México. [email protected] JORGE LLORENTE-BOUSQUETS Museo de Zoología, Facultad de Ciencias, UNAM. Apartado Postal 70-399, México D.F. 04510; México. [email protected]. Autor responsable. RESUMEN La riqueza biológica de Mesoamérica es enorme. Dentro de esta gran área geográfi ca se encuentran algunos de los ecosistemas más diversos del planeta (selvas tropicales), así como varios de los principales centros de endemismo en el mundo (bosques nublados). Países como Guatemala, en esta gran área biogeográfi ca, tiene grandes zonas de bosque húmedo tropical y bosque mesófi lo, por esta razón es muy importante para analizar la diversidad en la región. Lamentablemente, la fauna de mariposas de Guatemala es poco conocida y por lo tanto, es necesario llevar a cabo un estudio y análisis de la composición y la diversidad de las mariposas (Lepidoptera: Papilionoidea) en Guatemala. -

An Annotated Checklist of Ecuadorian Pieridae (Lepidoptera, Pieridae) 545-580 ©Ges
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Atalanta Jahr/Year: 1996 Band/Volume: 27 Autor(en)/Author(s): Racheli Tommaso Artikel/Article: An annotated checklist of Ecuadorian Pieridae (Lepidoptera, Pieridae) 545-580 ©Ges. zur Förderung d. Erforschung von Insektenwanderungen e.V. München, download unter www.zobodat.at Atalanta (December 1996) 27(3/4): 545-580, Wurzburg, ISSN 0171-0079 An annotated checklist of Ecuadorian Pieridae (Lepidoptera, Pieridae) by To m m a s o R a c h e li received 21.111.1996 Abstract: An account of 134 Pierid taxa occurring in Ecuador is presented. Data are from 12 years field experience in the country and from Museums specimens. Some new species records are added to Ecuadorian fauna and it is presumed that at least a 10% more of new records will be obtained in the near future. Ecuadorian Pieridae, although in the past many taxa were described from this country, are far from being thoroughly known. One of the most prolific author was Hewitson (1852-1877; 1869-1870; 1870; 1877) who described many species from the collections made by Buckley and Simons . Some of the "Ecuador” citations by Hewitson are pointed out more precisely by the same author (Hewit son , 1870) in his index to the list of species collected by Buckley in remote areas uneasily reached even to-day (V ane -Wright, 1991). An important contribution on Lepidoptera of Ecuador is given by Dognin (1887-1896) who described and listed many new species collected by Gaujon in the Loja area, where typical amazonian and páramo species are included. -

Combining Taxonomic and Functional Approaches to Unravel the Spatial Distribution of an Amazonian Butterfly Community
Environmental Entomology Advance Access published December 7, 2015 Environmental Entomology, 2015, 1–9 doi: 10.1093/ee/nvv183 Community and Ecosystem Ecology Research article Combining Taxonomic and Functional Approaches to Unravel the Spatial Distribution of an Amazonian Butterfly Community Marlon B. Grac¸a,1,2,3 Jose´W. Morais,1 Elizabeth Franklin,1,2 Pedro A. C. L. Pequeno,1,2 Jorge L. P. Souza,1,2 and Anderson Saldanha Bueno,1,4 1Biodiversity Coordination, National Institute for Amazonian Research, INPA, Manaus, Brazil ([email protected]; [email protected]; [email protected]; [email protected]; [email protected]; [email protected]), 2Center for Integrated Studies of Amazonian Biodiversity, CENBAM, Manaus, Brazil, 3Corresponding author, e-mail: marlon_lgp@hotmail. com, and 4Campus Ju´lio de Castilhos, Farroupilha Federal Institute of Education, Science and Technology, Brazil ([email protected]) Received 24 August 2015; Accepted 10 November 2015 Abstract This study investigated the spatial distribution of an Amazonian fruit-feeding butterfly assemblage by linking spe- cies taxonomic and functional approaches. We hypothesized that: 1) vegetation richness (i.e., resources) and abun- dance of insectivorous birds (i.e., predators) should drive changes in butterfly taxonomic composition, 2) larval diet breadth should decrease with increase of plant species richness, 3) small-sized adults should be favored by higher abundance of birds, and 4) communities with eyespot markings should be able to exploit areas with higher predation pressure. Fruit-feeding butterflies were sampled with bait traps and insect nets across 25 km2 of an Amazonian ombrophilous forest in Brazil. We measured larval diet breadth, adult body size, and wing marking of all butterflies. -

Extreme Diversity of Tropical Parasitoid Wasps Exposed by Iterative Integration of Natural History, DNA Barcoding, Morphology, and Collections
Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections M. Alex Smith*†, Josephine J. Rodriguez‡, James B. Whitfield‡, Andrew R. Deans§, Daniel H. Janzen†¶, Winnie Hallwachs¶, and Paul D. N. Hebert* *The Biodiversity Institute of Ontario, University of Guelph, Guelph Ontario, N1G 2W1 Canada; ‡Department of Entomology, 320 Morrill Hall, University of Illinois, 505 S. Goodwin Avenue, Urbana, IL 61801; §Department of Entomology, North Carolina State University, Campus Box 7613, 2301 Gardner Hall, Raleigh, NC 27695-7613; and ¶Department of Biology, University of Pennsylvania, Philadelphia, PA 19104-6018 Contributed by Daniel H. Janzen, May 31, 2008 (sent for review April 18, 2008) We DNA barcoded 2,597 parasitoid wasps belonging to 6 microgas- A detailed recognition of species in parasitoid communities is trine braconid genera reared from parapatric tropical dry forest, cloud necessary because of the pivotal role parasitoids play in food web forest, and rain forest in Area de Conservacio´ n Guanacaste (ACG) in structure and dynamics. While generalizations about the effects of northwestern Costa Rica and combined these data with records of parasitoids on community diversity are complex (7), a common- caterpillar hosts and morphological analyses. We asked whether place predictor of the impact of a parasitoid species on local host barcoding and morphology discover the same provisional species and dynamics is whether the parasitoid is a generalist or specialist. A whether the biological entities revealed by our analysis are congruent generalist, especially a mobile one, is viewed as stabilizing food webs with wasp host specificity. Morphological analysis revealed 171 (see ref. -
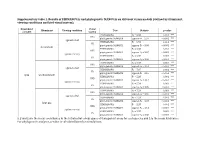
Supplementary Table 1. Results of Permanovas and Phylogenetic Manovas on Different Vision Models (Defined by Illuminant, Viewing Conditions and Bird Visual System)
Supplementary table 1. Results of PERMANOVAs and phylogenetic MANOVAs on different vision models (defined by illuminant, viewing conditions and bird visual system). Dependent Visual Illuminant Viewing condition Test Statistic p-value variable system PERMANOVA F9 = 6.88 0.001 *** UVS phylogenetic MANOVA approx-F9 = 2.97 < 0.001 *** against a leaf PERMANOVA F9 = 6.93 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.05 < 0.001 *** forest shade PERMANOVA F9 = 5.38 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.07 < 0.001 *** against the sky PERMANOVA F9 = 5.38 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.36 < 0.001 *** PERMANOVA F9 = 7.04 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.01 < 0.001 *** against a leaf PERMANOVA F9 = 7.07 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.10 < 0.001 *** xyzL woodland shade PERMANOVA F9 = 5.33 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.12 < 0.001 *** against the sky PERMANOVA F9 = 5.34 0.002 ** VS phylogenetic MANOVA approx-F9 = 3.39 < 0.001 *** PERMANOVA F9 = 7.24 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.00 < 0.001 *** against a leaf PERMANOVA F9 = 7.24 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.07 < 0.001 *** large gap PERMANOVA F9 = 5.37 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.14 < 0.001 *** against the sky PERMANOVA F9 = 5.37 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.38 < 0.001 *** x, y and z are the mean coordinates in the tetrahedral colour space of transparent areas for each species and L is the mean luminance. -

Revised Species Definitions and Nomenclature of the Rose Colored Cithaerias Butterflies (Lepidoptera, Nymphalidae, Satyrinae)
Zootaxa 3873 (5): 541–559 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2014 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3873.5.5 http://zoobank.org/urn:lsid:zoobank.org:pub:05BD334C-493D-4688-92E8-602943ECF57D Revised species definitions and nomenclature of the rose colored Cithaerias butterflies (Lepidoptera, Nymphalidae, Satyrinae) CARLA M. PENZ1, LAURA G. ALEXANDER2 & PHILIP J. DEVRIES3 Department of Biological Sciences, University of New Orleans, 2000 Lakeshore Dr. New Orleans, LA 70148, USA. E-mail: [email protected]; [email protected]; [email protected] Abstract This study provides updated species definitions for five rose-colored Cithaerias butterflies, starting with a historical over- view of their taxonomy. Given their mostly transparent wings, genitalia morphology yielded the most reliable characters for species definition and identification. Genitalic divergence is more pronounced when multiple species occur in sympa- try than between parapatric taxa. Cithaerias aurorina is granted full species status, C. cliftoni is reinstated as a full species, and one new combination is proposed, i.e. C. aurora tambopata. Two new synonyms are proposed, Callitaera phantoma and Callitaera aura = Cithaerias aurora. Key words: pireta, menander, aurorina, cliftoni, aurora, aura, phantoma, pyritosa Introduction Some of the most visually striking Neotropical butterflies belong to the genus Cithaerias Hübner (Satyrinae, Haeterini), which inhabit sea level to mid-elevation rainforests from Mexico through Central and South America. A characteristic of all Cithaerias species is their mostly transparent wings with the distal portions of the hind wing overlaid with partially lustrous rose, purple or blue scales. -

Differential Sequestration of a Cytotoxic Vismione from the Host Plant Vismia Baccifera by Periphoba Arcaei and Pyrrhopyge Thericles
JChemEcol DOI 10.1007/s10886-015-0614-6 Differential Sequestration of a Cytotoxic Vismione from the Host Plant Vismia baccifera by Periphoba arcaei and Pyrrhopyge thericles Ciara Raudsepp-Hearne 1,2,3 & Annette Aiello 1 & Ahmed A. Hussein4,5 & Maria V. Heller1,6 & Timothy Johns 2 & Todd L. Capson 1,2,7 Received: 6 May 2014 /Revised: 29 June 2015 /Accepted: 30 July 2015 # Springer Science+Business Media New York 2015 Abstract We sought to compare the abilities of the specialist fold greater than compound 1, indicating that the generalist Lepidoptera Pyrrhopyge thericles (Hesperiidae) and the gen- P. arcaei is capable of selectively sequestering cytotoxic com- eralist Periphoba arcaei (Saturniidae) to assimilate three high- pounds from its host plant. Compounds 1 and 2 show compa- ly cytotoxic compounds from their larval host plant, Vismia rable cytotoxicities in three different cancer cell lines, suggest- baccifera (Clusiaceae) and to determine whether either insect ing that properties other than cytotoxicity are responsible for discriminated in its assimilation of the compounds that are the selective sequestration of 1 by P. arcaei. This study repre- structurally similar but of variable cytotoxicity. Vismione B sents the first time that sequestration of this class of com- (1), deacetylvismione A (2), and deacetylvismione H (3)are pounds has been recorded in the Lepidoptera. cytotoxic compounds isolated from V.baccifera. Compound 1 was found in the 2nd and 3rd instars of P.arcaei,butnotinthe Keywords Cytotoxic . Sequestration . Aposematic . mature larvae or the pupae. Pyrrhopyge thericles assimilated Clusiaceae . Lepidoptera . Saturniidae . Hesperiidae trace quantities of compound 1 and deacetylvismione A (2), which were both found in the 3rd and 4th instars. -

NYMPHALIDAE: SATYRINAE Jaime Pinzón-C
WEB VERSION MARIPOSAS del Bajo Río Caquetá y Apaporis (Amazonia Colombiana) 1 NYMPHALIDAE: SATYRINAE Jaime Pinzón-C. - - Conservación Internacional Colombia Fotos de J. Pinzón-C. Producido por: T. S. Wachter, R. B. Foster, con el apoyo del Ellen Hyndman Fund y Andrew Mellon Foundation. © Jaime Pinzón-C. [[email protected]] Conservation International – Colombia, Cra 13 # 71-41 Bogotá, Colombia: [http://www.conservation.org.co] © Environmental & Conservation Programs, The Field Museum, Chicago, IL 60605 USA. [[email protected]] [www.fmnh.org/animalguides] Rapid Color Guide # 250 versión 1 02/2009 DORSAL VENTRAL Cithaerias pireta aurorina Cithaerias aff. pyritosa Haetera piera Bia actorion WEB VERSION 2 MARIPOSAS del Bajo Río Caquetá y Apaporis (Amazonia Colombiana) NYMPHALIDAE: SATYRINAE Jaime Pinzón-C. - - Conservación Internacional Colombia Fotos de J. Pinzón-C. Producido por: T. S. Wachter, R. B. Foster, con el apoyo del Ellen Hyndman Fund y Andrew Mellon Foundation. © Jaime Pinzón-C. [[email protected]] Conservation International – Colombia, Cra 13 # 71-41 Bogotá, Colombia: [http://www.conservation.org.co] © Environmental & Conservation Programs, The Field Museum, Chicago, IL 60605 USA. [[email protected]] [www.fmnh.org/animalguides] Rapid Color Guide # 250 versión 1 02/2009 DORSAL VENTRAL Pierella astyoche Pierella hortona Pierella lamia Pierella lena Caeruleuptychia caerulea Chloreuptychia agatha WEB VERSION MARIPOSAS del Bajo Río Caquetá y Apaporis (Amazonia Colombiana) 3 NYMPHALIDAE: SATYRINAE Jaime Pinzón-C. - - Conservación Internacional Colombia Fotos de J. Pinzón-C. Producido por: T. S. Wachter, R. B. Foster, con el apoyo del Ellen Hyndman Fund y Andrew Mellon Foundation. © Jaime Pinzón-C. [[email protected]] Conservation International – Colombia, Cra 13 # 71-41 Bogotá, Colombia: [http://www.conservation.org.co] © Environmental & Conservation Programs, The Field Museum, Chicago, IL 60605 USA.