Stervander Et Al. 2015 Mol Ecol
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Morocco SD 2017 Trip Report
Morocco 9th - 18th March 2017 Desert Sparrow is surely one of the best looking and most sought after of all the sparrows Tour Leader: Lisle Gwynn All photos in this report were taken by Lisle Gwynn on this tour Species depicted in photographs are named in BOLD RED www.tropicalbirding.com +1-409-515-9110 [email protected] Introduction Morocco is a fascinating destination, and one that many world birders have neglected for too long. It is increasingly becoming a go-to country for European birders in Spring, and offers some of the most exciting birding in the Western Palearctic biogeographic region. Not only does it offer a chance to see Afro-European migration at its peak, but it also offers a plethora of exciting and special endemic and near-endemic species at its core. Add to this the fact that throughout the tour we have excellent accommodation and some of the best food available anywhere in the world (in my opinion), it all goes toward making Morocco a must-visit location for any birder branching out into the world. It is also currently by far the safest North African country to visit, with little crime and none of the problems that plague the rest of the region, and therefore presents a comfortable and safe opportunity to experience North Africa. This year’s tour followed our tried and tested route, starting in the manic city of Marrakesh at a serene hotel amongst the craziness, a quick departure to the idyllic Ourika Valley and the high snow-capped peaks of Oukameiden and the high Atlas Mountains, before descending to the stony desert around Boumalne Dades and the ochre-cast dunes of the Sahara at Erg Chebbi. -

American Robin
American Robin DuPage Birding Club, 2020 American Robin Appearance A chunky, heavy-bodied bird with a relatively small dark head. Sexually dimorphic, meaning the male and female look different. American Robins are a uniform dark gray with a brick red breast. Female Male Females are a lighter gray with a lighter breast. Males tend to be darker with a brighter red breast. Males are larger than females. Photos: Elmarie Von Rooyen (left), Jackie Tilles (right) DuPage Birding Club, 2020 2 American Robin Appearance American Robins are a medium-size bird with a length of about ten inches. They are so common that they are a good bird to compare size with when you come across an unknown bird. Is the bird bigger than an American Robin or smaller than an American Robin? Judging the size of a bird is very helpful in identifying an unknown bird. Chart: The Cornell Lab, All About Birds https://www.allaboutbirds.org/guide/American_Robin/id DuPage Birding Club, 2020 3 American Robin Appearance Juvenile American Robins have a speckled breast with a tint of rusty red. Photos: Natalie McFaul DuPage Birding Club, 2020 4 American Robin Sounds From The Cornell Lab of Ornithology: https://www.birds.cornell.edu/home/ SONGS The musical song of the American Robin is a familiar sound of spring. It’s a string of 10 or so clear whistles assembled from a few often- repeated syllables, and often described as cheerily, cheer up, cheer up, cheerily, cheer up. The syllables rise and fall in pitch but are delivered at a steady rhythm, with a pause before the bird begins singing again. -

Implications of Conspecific Background Noise for Features of Blue Tit, Cyanistes Caeruleus, Communication Networks at Dawn
J Ornithol (2007) 148:123–128 DOI 10.1007/s10336-006-0116-y ORIGINAL ARTICLE Implications of conspecific background noise for features of blue tit, Cyanistes caeruleus, communication networks at dawn Angelika Poesel Æ Torben Dabelsteen Æ Simon Boel Pedersen Received: 3 January 2006 / Revised: 8 November 2006 / Accepted: 8 November 2006 / Published online: 5 December 2006 Ó Dt. Ornithologen-Gesellschaft e.V. 2006 Abstract Communication among animals often com- may potentially constrain the perception of singing prises several signallers and receivers within the sig- patterns and may constitute costs for eavesdroppers. nal’s transmission range. In such communication On the other hand, signallers may position themselves networks, individuals can extract information about strategically and privatise their interactions. differences in relative performance of conspecifics by eavesdropping on their signalling interactions. In Keywords Communication network Á Cyanistes songbirds, information can be encoded in the timing of caeruleus Á Singing interaction signals, which either alternate or overlap, and both male and female receivers may utilise this information when engaging in territorial interactions or making Introduction reproductive decisions, respectively. We investigated how conspecific background noise at dawn may overlay Territorial animals form neighbourhoods of several and possibly constrain the perception of such singing individuals within signalling and receiving range (see patterns. We simulated a small communication net- examples in McGregor 2005). In order to defend ter- work of blue tits, Cyanistes caeruleus, at dawn in ritorial space successfully, it may be advantageous to spring. Two loudspeakers simulated a singing interac- gather information about competitors and thus be able tion which was recorded from four different receiver to adjust responses to an opponent’s strength. -

Thése REBBAH Abderraouf Chouaib Bibliothéque.Pdf
République Algérienne Démocratique et Populaire Ministère de l’Enseignement Supérieur et de la Recherche Scientifique Université Larbi Ben M’hidi Oum El Bouaghi Faculté Des Sciences Exactes et des Sciences de la Nature et de la Vie Département des Sciences de la Nature et de la Vie Thèse Présentée en vue de l’obtention du diplôme Doctorat LMD en Sciences de la nature Option: Structure et dynamique des écosystèmes Théme INVENTAIRE ET ECOLOGIE DES OISEAUX FORESTIERS DE DJEBEL SIDI REGHIS (OUM EL BOUAGHI) Présentée par : Mr.REBBAH Abderraouf Chouaib Membres du Jury: Président: BELAIDI Abdelhakim Pr (Université Larbi Ben Mhidi, Oum El-Bouaghi). Promoteur : SAHEBMenouar Pr (Université Larbi Ben Mhidi, Oum El-Bouaghi). Examinateurs: ABABSA Labed Pr (Université Larbi Ben Mhidi, Oum El-Bouaghi). Examinateurs: HOUHAMDI Moussa Pr (Université de Guelma). Examinateurs: OUAKID Mohamed Laid Pr (Université d’Annaba). Année universitaire: 2018-2019 << ِ ِ أَﻟَْﻢ ﺗَ َﺮ أَ ﱠن ﱠاﻪﻠﻟَ ﻳُﺴَﺒِّ ُﺢ ﻟَﻪُ ﻣَ ْﻦ ﻓﻲ اﻟﺴﱠﻤَ َﺎوات َو ْاﻷَ ْر ِض َواﻟﻄﱠْﻴ ُﺮ ٍ ۖ◌ ِ ِ ۗ◌ ِ ِ ﺻَ ﺎ ﻓ ـﱠ ﺎ ت ُﻛ ﻞﱞ ﻗَ ْﺪ ﻋَ ﻠ ﻢَ ﺻَ َﻼ ﺗَ ﻪُ َو ﺗَ ْﺴ ﺒ ﻴ ﺤَ ﻪُ َو ﱠاﻪﻠﻟُ ﻋَﻠﻴﻢٌ ﺑﻤَﺎ ﻳَﻔْ َﻌﻠُ َﻮن >> ﺳﻮرة اﻟﻨﻮراﻷﻳﺔ 41 Dédicaces Je dédie ce travail à : A mes parents qui m’ont tout donné, et qui étaient toujours la à coté de moi dans chaque pats depuis le premier crie pour m’aidé, m’orienté avec leurs amour et leurs sacrifices, malgré les couts dures de la vie. Aucun hommage ne pourrait être à la hauteur de l’amour Dont ils ne cessent de me combler. -

Progress in the Development of an Eurasian-African Bird Migration Atlas
CONVENTION ON UNEP/CMS/COP13/Inf.20 MIGRATORY 10 February 2020 SPECIES Original: English 13th MEETING OF THE CONFERENCE OF THE PARTIES Gandhinagar, India, 17 - 22 February 2020 Agenda Item 25 PROGRESS IN THE DEVELOPMENT OF AN EURASIAN-AFRICAN BIRD MIGRATION ATLAS (Submitted by the European Union of Bird Ringing (EURING) and the Institute of Avian Research) Summary: The African-Eurasian Bird Migration Atlas is being developed under the auspices of CMS in the framework of a Global Animal Migration Atlas, of which it constitutes a module. The African-Eurasian Bird Migration Atlas is being developed and compiled by the European Union of Bird Ringing (EURING) under a Project Cooperation Agreement (PCA) between the CMS Secretariat and the Institute of Avian Research, acting on behalf of EURING. The development of the African-Eurasian Bird Migration Atlas is funded with the contribution granted by the Government of Italy under the Migratory Species Champion Programme. This information document includes a progress report on the development of the various components of the project. The project is expected to be completed in 2021. UNEP/CMS/COP13/Inf.20 Eurasian-African Bird Migration Atlas progress report February 2020 Stephen Baillie1, Franz Bairlein2, Wolfgang Fiedler3, Fernando Spina4, Kasper Thorup5, Sam Franks1, Dorian Moss1, Justin Walker1, Daniel Higgins1, Roberto Ambrosini6, Niccolò Fattorini6, Juan Arizaga7, Maite Laso7, Frédéric Jiguet8, Boris Nikolov9, Henk van der Jeugd10, Andy Musgrove1, Mark Hammond1 and William Skellorn1. A report to the Convention on Migratory Species from the European Union for Bird Ringing (EURING) and the Institite of Avian Research, Wilhelmshaven, Germany 1. British Trust for Ornithology, Thetford, IP24 2PU, UK 2. -

Territoriality As a Paternity Guard in the European Robin, Erithacus Rubecula
ANIMAL BEHAVIOUR, 2000, 60, 165–173 doi:10.1006/anbe.2000.1442, available online at http://www.idealibrary.com on ARTICLES Territoriality as a paternity guard in the European robin, Erithacus rubecula JOE TOBIAS & NAT SEDDON Department of Zoology, University of Cambridge (Received 29 June 1999; initial acceptance 29 July 1999; final acceptance 18 March 2000; MS. number: 6273) To investigate the relative importance of paternity defences in the European robin we used behavioural observations, simulated intrusions and temporary male removal experiments. Given that paired males did not increase their mate attendance, copulation rate or territory size during the female’s fertile period, the most frequently quoted paternity assurance strategies in birds were absent. However, males with fertile females sang and patrolled their territories more regularly, suggesting that territorial motivation and vigilance were elevated when the risk of cuckoldry was greatest. In addition, there was a significant effect of breeding period on response to simulated intrusions: residents approached and attacked freeze-dried mounts more readily in the fertile period. During 90-min removals of the pair male in the fertile period, neighbours trespassed more frequently relative to prefertile and fertile period controls and appeared to seek copulations with unattended females. When replaced on their territories, males immediately increased both song rate and patrolling rate in comparison with controls. We propose that male robins sing to signal their presence, and increase their territorial vigilance and aggression in the fertile period to protect paternity. 2000 The Association for the Study of Animal Behaviour Despite the apparent social monogamy of the majority of Thryothorus nigricapillus: Levin 1996). -
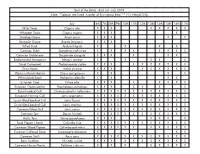
Best of the Baltic - Bird List - July 2019 Note: *Species Are Listed in Order of First Seeing Them ** H = Heard Only
Best of the Baltic - Bird List - July 2019 Note: *Species are listed in order of first seeing them ** H = Heard Only July 6th 7th 8th 9th 10th 11th 12th 13th 14th 15th 16th 17th Mute Swan Cygnus olor X X X X X X X X Whopper Swan Cygnus cygnus X X X X Greylag Goose Anser anser X X X X X Barnacle Goose Branta leucopsis X X X Tufted Duck Aythya fuligula X X X X Common Eider Somateria mollissima X X X X X X X X Common Goldeneye Bucephala clangula X X X X X X Red-breasted Merganser Mergus serrator X X X X X Great Cormorant Phalacrocorax carbo X X X X X X X X X X Grey Heron Ardea cinerea X X X X X X X X X Western Marsh Harrier Circus aeruginosus X X X X White-tailed Eagle Haliaeetus albicilla X X X X Eurasian Coot Fulica atra X X X X X X X X Eurasian Oystercatcher Haematopus ostralegus X X X X X X X Black-headed Gull Chroicocephalus ridibundus X X X X X X X X X X X X European Herring Gull Larus argentatus X X X X X X X X X X X X Lesser Black-backed Gull Larus fuscus X X X X X X X X X X X X Great Black-backed Gull Larus marinus X X X X X X X X X X X X Common/Mew Gull Larus canus X X X X X X X X X X X X Common Tern Sterna hirundo X X X X X X X X X X X X Arctic Tern Sterna paradisaea X X X X X X X Feral Pigeon ( Rock) Columba livia X X X X X X X X X X X X Common Wood Pigeon Columba palumbus X X X X X X X X X X X Eurasian Collared Dove Streptopelia decaocto X X X Common Swift Apus apus X X X X X X X X X X X X Barn Swallow Hirundo rustica X X X X X X X X X X X Common House Martin Delichon urbicum X X X X X X X X White Wagtail Motacilla alba X X -

United Nations Educational, Scientific and Cultural Organization
SC-20/CONF.232/8 Paris, 27 September 2020 Original: English UNITED NATIONS EDUCATIONAL, SCIENTIFIC AND CULTURAL ORGANIZATION International Co-ordinating Council of the Man and the Biosphere (MAB) Programme Thirty-second session Online session 27– 28 September 2020 ITEM 9 OF THE PROVISIONAL AGENDA: Proposals for New Biosphere Reserves and Extensions/ Modifications/ Renaming to Biosphere Reserves that are part of the World Network of Biosphere Reserves (WNBR) The Secretariat of the United Nations Educational Scientific and Cultural Organization (UNESCO) does not represent or endorse the accuracy or reliability of any advice, opinion, statement or other information or documentation provided by States to the Secretariat of UNESCO. The publication of any such advice, opinion, statement or other information or documentation on UNESCO’s website and/or on working documents also does not imply the expression of any opinion whatsoever on the part of the Secretariat of UNESCO concerning the legal status of any country, territory, city or area or of its boundaries. 1. Proposals for new biosphere reserves and extensions to biosphere reserves that are already part of the World Network of Biosphere Reserves (WNBR) were considered at the 26th meeting of the International Advisory Committee for Biosphere Reserves (IACBR), which met at UNESCO Headquarters from 17 to 20 February 2020. 2. The members of the Advisory Committee examined 30 proposals: 25 for new biosphere reserves including one transboundary site, two resubmissions and three requests for extensions/modification and/or renaming of already existing biosphere reserves. Five new counries submitted proposals: Andora, Cap verde, Comoros, Luxemburg an Trinidad and Tobago. -

Habitat Modelling and the Ecology of the Marsh Tit (Poecile Palustris)
HABITAT MODELLING AND THE ECOLOGY OF THE MARSH TIT (POECILE PALUSTRIS) RICHARD K BROUGHTON A thesis submitted in partial fulfilment of the requirements of Bournemouth University for the degree of Doctor of Philosophy August 2012 Bournemouth University in collaboration with the Centre for Ecology & Hydrology This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with its author and due acknowledgement must always be made of the use of any material contained in, or derived from, this thesis. 2 ABSTRACT Richard K Broughton Habitat modelling and the ecology of the Marsh Tit (Poecile palustris) Among British birds, a number of woodland specialists have undergone a serious population decline in recent decades, for reasons that are poorly understood. The Marsh Tit is one such species, experiencing a 71% decline in abundance between 1967 and 2009, and a 17% range contraction between 1968 and 1991. The factors driving this decline are uncertain, but hypotheses include a reduction in breeding success and annual survival, increased inter-specific competition, and deteriorating habitat quality. Despite recent work investigating some of these elements, knowledge of the Marsh Tit’s behaviour, landscape ecology and habitat selection remains incomplete, limiting the understanding of the species’ decline. This thesis provides additional key information on the ecology of the Marsh Tit with which to test and review leading hypotheses for the species’ decline. Using novel analytical methods, comprehensive high-resolution models of woodland habitat derived from airborne remote sensing were combined with extensive datasets of Marsh Tit territory and nest-site locations to describe habitat selection in unprecedented detail. -

Sparrow Swap: Testing Management Strategies for House Sparrows and Exploring the Use of Their Eggshells for Monitoring Heavy Metal Pollution
ABSTRACT HARTLEY, SUZANNE MARIE. Sparrow Swap: Testing Management Strategies for House Sparrows and Exploring the Use of their Eggshells for Monitoring Heavy Metal Pollution. (Under the direction of Dr. Caren Cooper). Human movement across the globe, particularly through colonialism throughout the last 500 years, has led to the introduction of species into novel environments where they threaten the biodiversity and ecosystem functioning of those novel environments. In the Anthropocene where other threats such as climate change, pollution, and habitat destruction already occur, invasive species are just one more threat facing ecosystems. But what if we can find a way to use an invasive species to help monitor those other threats while at the same time managing them? In the following thesis I explore the strategies by which volunteers manage House Sparrows to minimize their negative impact as an invasive species, but also the potential to use their eggs as indicators of heavy metals in the environment. House Sparrows compete with native birds for nesting spaces. They are also commensal with humans, utilizing buildings as nesting spaces and split grains and forgotten French fries as food sources. In order to 1) find effective management strategies for House Sparrows and 2) evaluate their use as indicators of environmental contaminants, a citizen science project Sparrow Swap was created. Sparrow Swaps takes advantage of the ubiquity of House Sparrows and the expertise of volunteer nestbox monitors to gather data about House Sparrow nesting behaviors and eggs across the United States. In Chapter 1, I address the first research goal of Sparrow Swap by comparing the outcomes of two different strategies by which volunteers manage House Sparrows. -

Egg Recognition in Cinereous Tits (Parus Cinereus): Eggshell Spots Matter Jianping Liu1 , Canchao Yang1 , Jiangping Yu2,3 , Haitao Wang2,4 and Wei Liang1*
Liu et al. Avian Res (2019) 10:37 https://doi.org/10.1186/s40657-019-0178-1 Avian Research RESEARCH Open Access Egg recognition in Cinereous Tits (Parus cinereus): eggshell spots matter Jianping Liu1 , Canchao Yang1 , Jiangping Yu2,3 , Haitao Wang2,4 and Wei Liang1* Abstract Background: Brood parasitic birds such as cuckoos (Cuculus spp.) can reduce their host’s reproductive success. Such selection pressure on the hosts has driven the evolution of defense behaviors such as egg rejection against cuckoo parasitism. Studies have shown that Cinereous Tits (Parus cinereus) in China have a good ability for recognizing foreign eggs. However, it is unclear whether egg spots play a role in egg recognition. The aims of our study were to inves- tigate the egg recognition ability of two Cinereous Tit populations in China and to explore the role of spots in egg recognition. Methods: To test the efect of eggshell spots on egg recognition, pure white eggs of the White-rumped Munia (Lon- chura striata) and eggs of White-rumped Munia painted with red brown spots were used to simulate experimental parasitism. Results: Egg experiments showed that Cinereous Tits rejected 51.5% of pure white eggs of the White-rumped Munia, but only 14.3% of spotted eggs of the White-rumped Munia. There was a signifcant diference in egg recognition and rejection rate between the two egg types. Conclusions: We conclude that eggshell spots on Cinereous Tit eggs had a signaling function and may be essential to tits for recognizing and rejecting parasitic eggs. Keywords: Brood parasitism, Egg recognition, Egg rejection, Eggshell spots, Parus cinereus Background egg rejection by hosts, many parasitic birds evolve coun- Te mutual adaptations and counter-defense strategies ter-adaptations to overcome the hosts’ defenses by laying between brood parasitic birds such as cuckoos (Cuculus mimicking (Brooke and Davies 1988; Avilés et al. -

High Level of Self-Control Ability in a Small Passerine Bird
Behavioral Ecology and Sociobiology (2018) 72: 118 https://doi.org/10.1007/s00265-018-2529-z ORIGINAL ARTICLE High level of self-control ability in a small passerine bird Emil Isaksson1 & A. Utku Urhan1 & Anders Brodin1 Received: 27 October 2017 /Revised: 8 June 2018 /Accepted: 14 June 2018 /Published online: 26 June 2018 # The Author(s) 2018 Abstract Cognitively advanced animals are usually assumed to possess better self-control, or ability to decline immediate rewards in favour of delayed ones, than less cognitively advanced animals. It has been claimed that the best predictor of high such ability is absolute brain volume meaning that large-brained animals should perform better than small-brained ones. We tested self-control ability in the great tit, a small passerine. In the common test of this ability, the animal is presented with a transparent cylinder that contains a piece of food. If the animal tries to take the reward through the transparent wall of the cylinder, this is considered an impulsive act and it fails the test. If it moves to an opening and takes the reward this way, it passes the test. The average performance of our great tits was 80%, higher than most animals that have been tested and almost in level with the performance in corvids and apes. This is remarkable considering that the brain volume of a great tit is 3% of that of a raven and 0.1% of that of a chimpanzee. Significance statement The transparent cylinder test is the most common way to test the ability of self-control in animals.