Comparing Detectability Patterns of Bird Species in Small Ponds Using
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Disaggregation of Bird Families Listed on Cms Appendix Ii
Convention on the Conservation of Migratory Species of Wild Animals 2nd Meeting of the Sessional Committee of the CMS Scientific Council (ScC-SC2) Bonn, Germany, 10 – 14 July 2017 UNEP/CMS/ScC-SC2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II (Prepared by the Appointed Councillors for Birds) Summary: The first meeting of the Sessional Committee of the Scientific Council identified the adoption of a new standard reference for avian taxonomy as an opportunity to disaggregate the higher-level taxa listed on Appendix II and to identify those that are considered to be migratory species and that have an unfavourable conservation status. The current paper presents an initial analysis of the higher-level disaggregation using the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World Volumes 1 and 2 taxonomy, and identifies the challenges in completing the analysis to identify all of the migratory species and the corresponding Range States. The document has been prepared by the COP Appointed Scientific Councilors for Birds. This is a supplementary paper to COP document UNEP/CMS/COP12/Doc.25.3 on Taxonomy and Nomenclature UNEP/CMS/ScC-Sc2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II 1. Through Resolution 11.19, the Conference of Parties adopted as the standard reference for bird taxonomy and nomenclature for Non-Passerine species the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World, Volume 1: Non-Passerines, by Josep del Hoyo and Nigel J. Collar (2014); 2. -
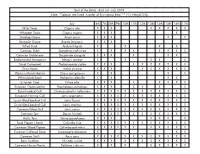
Best of the Baltic - Bird List - July 2019 Note: *Species Are Listed in Order of First Seeing Them ** H = Heard Only
Best of the Baltic - Bird List - July 2019 Note: *Species are listed in order of first seeing them ** H = Heard Only July 6th 7th 8th 9th 10th 11th 12th 13th 14th 15th 16th 17th Mute Swan Cygnus olor X X X X X X X X Whopper Swan Cygnus cygnus X X X X Greylag Goose Anser anser X X X X X Barnacle Goose Branta leucopsis X X X Tufted Duck Aythya fuligula X X X X Common Eider Somateria mollissima X X X X X X X X Common Goldeneye Bucephala clangula X X X X X X Red-breasted Merganser Mergus serrator X X X X X Great Cormorant Phalacrocorax carbo X X X X X X X X X X Grey Heron Ardea cinerea X X X X X X X X X Western Marsh Harrier Circus aeruginosus X X X X White-tailed Eagle Haliaeetus albicilla X X X X Eurasian Coot Fulica atra X X X X X X X X Eurasian Oystercatcher Haematopus ostralegus X X X X X X X Black-headed Gull Chroicocephalus ridibundus X X X X X X X X X X X X European Herring Gull Larus argentatus X X X X X X X X X X X X Lesser Black-backed Gull Larus fuscus X X X X X X X X X X X X Great Black-backed Gull Larus marinus X X X X X X X X X X X X Common/Mew Gull Larus canus X X X X X X X X X X X X Common Tern Sterna hirundo X X X X X X X X X X X X Arctic Tern Sterna paradisaea X X X X X X X Feral Pigeon ( Rock) Columba livia X X X X X X X X X X X X Common Wood Pigeon Columba palumbus X X X X X X X X X X X Eurasian Collared Dove Streptopelia decaocto X X X Common Swift Apus apus X X X X X X X X X X X X Barn Swallow Hirundo rustica X X X X X X X X X X X Common House Martin Delichon urbicum X X X X X X X X White Wagtail Motacilla alba X X -

Discovery of a Relict Lineage and Monotypic Family of Passerine Birds
Discovery of a relict lineage and monotypic family of passerine birds Based on a comprehensive molecular dataset of passerines birds we identified a branch with a single species, the Spotted Wren-babbler Spelaeornis formosus. We suggest that this represents a relict lineage, which we propose should be placed in its own family, Elachuridae. The scientific name Elachura formosa should be used. We analysed of one of the most comprehensive datasets to date of the largest passerine bird clade, Passerida, which comprises c. 36% of the World’s c. 10,500 bird species. We identified 10 primary branches in the tree. One of these primary branches was made up of a single species, the Spotted Wren-Babbler Spelaeornis formosus, which is a small Wren-like bird that occurs in mountains from the eastern Himalayas to southeast China. This species apparently represents an old branch in the large passerine tree, without any close living relatives. There have surely been other relatives on this branch, which have gone extinct. The fact that it resembles wren-babblers and wrens in appearance is either due to pure chance or to convergent evolution, which may result in similar appearances in unrelated species that live in similar environments. We proposed the new family name Elachuridae for this single species. We also suggested that the scientific name Elachura formosa should be used, and the English name be changed to Elachura, to highlight its distinctness. Timaliidae (56) Pellorneidae (69) Leiothrichidae (133) Zosteropidae (128) Sylviidae (70) Pnoepygidae -

Common Birds of the Estero Bay Area
Common Birds of the Estero Bay Area Jeremy Beaulieu Lisa Andreano Michael Walgren Introduction The following is a guide to the common birds of the Estero Bay Area. Brief descriptions are provided as well as active months and status listings. Photos are primarily courtesy of Greg Smith. Species are arranged by family according to the Sibley Guide to Birds (2000). Gaviidae Red-throated Loon Gavia stellata Occurrence: Common Active Months: November-April Federal Status: None State/Audubon Status: None Description: A small loon seldom seen far from salt water. In the non-breeding season they have a grey face and red throat. They have a long slender dark bill and white speckling on their dark back. Information: These birds are winter residents to the Central Coast. Wintering Red- throated Loons can gather in large numbers in Morro Bay if food is abundant. They are common on salt water of all depths but frequently forage in shallow bays and estuaries rather than far out at sea. Because their legs are located so far back, loons have difficulty walking on land and are rarely found far from water. Most loons must paddle furiously across the surface of the water before becoming airborne, but these small loons can practically spring directly into the air from land, a useful ability on its artic tundra breeding grounds. Pacific Loon Gavia pacifica Occurrence: Common Active Months: November-April Federal Status: None State/Audubon Status: None Description: The Pacific Loon has a shorter neck than the Red-throated Loon. The bill is very straight and the head is very smoothly rounded. -

Foraging Behavior of Plain-Mantled Tit-Spinetail (Leptasthenura Aegithaloides) in Semiarid Matorral, North-Central Chile
ORNITOLOGIA NEOTROPICAL 22: 247–256, 2011 © The Neotropical Ornithological Society FORAGING BEHAVIOR OF PLAIN-MANTLED TIT-SPINETAIL (LEPTASTHENURA AEGITHALOIDES) IN SEMIARID MATORRAL, NORTH-CENTRAL CHILE Andrew Engilis Jr. & Douglas A. Kelt Department of Wildlife, Fish, and Conservation Biology - University of California, One Shields Avenue, Davis, CA 95616, USA. E-mail: [email protected] Resumen. – Comportamiento de forrajeo del tijeral (Leptasthenura aegithaloides) en matorral semiárido, centro-norte de Chile. – Hemos estudiado el comportamiento de forrajeo del tijeral (Leptas- thenura aegithaloides) en el matorral del centro-norte de Chile. Se trata de una especie de la familia Fur- nariidae que es insectívora recolectora desde perchas. Frecuenta los arbustos más dominantes y busca presas y alimentos principalmente en el follaje, grupos de flores, pequeñas ramas y masas de líquenes. Los arbustos preferidos incluyen a Porlieria y Baccharis. Se alimentan desde alturas cercanas al suelo hasta arbustos superiores a dos metros de altura. Se los encuentra más frecuentemente en parejas o en grupos pequeños, posiblemente familias, de tres a cinco aves. Las densidades promedio en el matorral (1,49 - 1,69 aves por hectárea) son mayores que las reportadas para otros lugares. Los tijerales en el matorral forman grupos de especies mixtas con facilidad, especialmente en el invierno Austral. Su estrategia de forrajeo y su comportamiento son similares a las del Mito sastrecillo de América del Norte (Psaltriparus minimus) y del Mito común (Aegithalos caudatus), ambos de la familia Aegithalidae, sugir- iendo estrategias ecológicas convergentes en ambientes estructuralmente similares. Abstract. – We studied foraging behavior of Plain-mantled Tit-spinetail (Leptasthenura aegithaloides) in matorral (scrubland) habitat of north-central Chile. -

Gear for a Big Year
APPENDIX 1 GEAR FOR A BIG YEAR 40-liter REI Vagabond Tour 40 Two passports Travel Pack Wallet Tumi luggage tag Two notebooks Leica 10x42 Ultravid HD-Plus Two Sharpie pens binoculars Oakley sunglasses Leica 65 mm Televid spotting scope with tripod Fossil watch Leica V-Lux camera Asics GEL-Enduro 7 trail running shoes GoPro Hero3 video camera with selfie stick Four Mountain Hardwear Wicked Lite short-sleeved T-shirts 11” MacBook Air laptop Columbia Sportswear rain shell iPhone 6 (and iPhone 4) with an international phone plan Marmot down jacket iPod nano and headphones Two pairs of ExOfficio field pants SureFire Fury LED flashlight Three pairs of ExOfficio Give- with rechargeable batteries N-Go boxer underwear Green laser pointer Two long-sleeved ExOfficio BugsAway insect-repelling Yalumi LED headlamp shirts with sun protection Sea to Summit silk sleeping bag Two pairs of SmartWool socks liner Two pairs of cotton Balega socks Set of adapter plugs for the world Birding Without Borders_F.indd 264 7/14/17 10:49 AM Gear for a Big Year • 265 Wildy Adventure anti-leech Antimalarial pills socks First-aid kit Two bandanas Assorted toiletries (comb, Plain black baseball cap lip balm, eye drops, toenail clippers, tweezers, toothbrush, REI Campware spoon toothpaste, floss, aspirin, Israeli water-purification tablets Imodium, sunscreen) Birding Without Borders_F.indd 265 7/14/17 10:49 AM APPENDIX 2 BIG YEAR SNAPSHOT New Unique per per % % Country Days Total New Unique Day Day New Unique Antarctica / Falklands 8 54 54 30 7 4 100% 56% Argentina 12 435 -

Niche Analysis and Conservation of Bird Species Using Urban Core Areas
sustainability Article Niche Analysis and Conservation of Bird Species Using Urban Core Areas Vasilios Liordos 1,* , Jukka Jokimäki 2 , Marja-Liisa Kaisanlahti-Jokimäki 2, Evangelos Valsamidis 1 and Vasileios J. Kontsiotis 1 1 Department of Forest and Natural Environment Sciences, International Hellenic University, 66100 Drama, Greece; [email protected] (E.V.); [email protected] (V.J.K.) 2 Arctic Centre, University of Lapland, 96101 Rovaniemi, Finland; jukka.jokimaki@ulapland.fi (J.J.); marja-liisa.kaisanlahti@ulapland.fi (M.-L.K.-J.) * Correspondence: [email protected] Abstract: Knowing the ecological requirements of bird species is essential for their successful con- servation. We studied the niche characteristics of birds in managed small-sized green spaces in the urban core areas of southern (Kavala, Greece) and northern Europe (Rovaniemi, Finland), during the breeding season, based on a set of 16 environmental variables and using Outlying Mean Index, a multivariate ordination technique. Overall, 26 bird species in Kavala and 15 in Rovaniemi were recorded in more than 5% of the green spaces and were used in detailed analyses. In both areas, bird species occupied different niches of varying marginality and breadth, indicating varying responses to urban environmental conditions. Birds showed high specialization in niche position, with 12 species in Kavala (46.2%) and six species in Rovaniemi (40.0%) having marginal niches. Niche breadth was narrower in Rovaniemi than in Kavala. Species in both communities were more strongly associated either with large green spaces located further away from the city center and having a high vegetation cover (urban adapters; e.g., Common Chaffinch (Fringilla coelebs), European Greenfinch (Chloris Citation: Liordos, V.; Jokimäki, J.; chloris Cyanistes caeruleus Kaisanlahti-Jokimäki, M.-L.; ), Eurasian Blue Tit ( )) or with green spaces located closer to the city center Valsamidis, E.; Kontsiotis, V.J. -

Federal Register/Vol. 85, No. 74/Thursday, April 16, 2020/Notices
21262 Federal Register / Vol. 85, No. 74 / Thursday, April 16, 2020 / Notices acquisition were not included in the 5275 Leesburg Pike, Falls Church, VA Comment (1): We received one calculation for TDC, the TDC limit would not 22041–3803; (703) 358–2376. comment from the Western Energy have exceeded amongst other items. SUPPLEMENTARY INFORMATION: Alliance, which requested that we Contact: Robert E. Mulderig, Deputy include European starling (Sturnus Assistant Secretary, Office of Public Housing What is the purpose of this notice? vulgaris) and house sparrow (Passer Investments, Office of Public and Indian Housing, Department of Housing and Urban The purpose of this notice is to domesticus) on the list of bird species Development, 451 Seventh Street SW, Room provide the public an updated list of not protected by the MBTA. 4130, Washington, DC 20410, telephone (202) ‘‘all nonnative, human-introduced bird Response: The draft list of nonnative, 402–4780. species to which the Migratory Bird human-introduced species was [FR Doc. 2020–08052 Filed 4–15–20; 8:45 am]‘ Treaty Act (16 U.S.C. 703 et seq.) does restricted to species belonging to biological families of migratory birds BILLING CODE 4210–67–P not apply,’’ as described in the MBTRA of 2004 (Division E, Title I, Sec. 143 of covered under any of the migratory bird the Consolidated Appropriations Act, treaties with Great Britain (for Canada), Mexico, Russia, or Japan. We excluded DEPARTMENT OF THE INTERIOR 2005; Pub. L. 108–447). The MBTRA states that ‘‘[a]s necessary, the Secretary species not occurring in biological Fish and Wildlife Service may update and publish the list of families included in the treaties from species exempted from protection of the the draft list. -

Comparative Analysis of Hissing Calls in Five Tit Species Li Zhang, Jianping Liu, Zezhong Gao, Lei Zhang, Dongmei Wan, Wei Liang, Anders Pape Møller
Comparative analysis of hissing calls in five tit species Li Zhang, Jianping Liu, Zezhong Gao, Lei Zhang, Dongmei Wan, Wei Liang, Anders Pape Møller To cite this version: Li Zhang, Jianping Liu, Zezhong Gao, Lei Zhang, Dongmei Wan, et al.. Comparative analysis of hissing calls in five tit species. Behavioural Processes, Elsevier, 2019. hal-03024634 HAL Id: hal-03024634 https://hal-cnrs.archives-ouvertes.fr/hal-03024634 Submitted on 25 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 Comparative analysis of hissing calls in five tit species 1 2 1 1 1, * 2 Li Zhang , Jianping Liu , Zezhong Gao , Lei Zhang , Dongmei Wan , Wei 2, * 3 3 Liang , Anders Pape Møller 1 4 Key Laboratory of Animal Resource and Epidemic Disease Prevention, 5 College of Life Sciences, Liaoning University, Shenyang 110036, China 2 6 Ministry of Education Key Laboratory for Ecology of Tropical Islands, 7 College of Life Sciences, Hainan Normal University, Haikou 571158, China 3 8 Ecologie Systématique Evolution, Université Paris-Sud, CNRS, 9 AgroParisTech, Université Paris-Saclay, F-91405 Orsay Cedex, France 10 Email address: 11 LZ, [email protected], ID: 0000-0003-1224-7855 12 JL, [email protected], ID: 0000-0001-6526-8831 13 ZG, [email protected] 14 LZ, [email protected], ID: 0000-0003-2328-1194 15 DW, [email protected], ID: 0000-0002-1465-6110 16 WL, [email protected], ID: 0000-0002-0004-9707 17 APM, [email protected], ID: 0000-0003-3739-4675 18 Word count: 4571 19 *Corresponding author. -

Supplementary Information For
Supplementary Information for Earth history and the passerine superradiation Oliveros, Carl H., Daniel J. Field, Daniel T. Ksepka, F. Keith Barker, Alexandre Aleixo, Michael J. Andersen, Per Alström, Brett W. Benz, Edward L. Braun, Michael J. Braun, Gustavo A. Bravo, Robb T. Brumfield, R. Terry Chesser, Santiago Claramunt, Joel Cracraft, Andrés M. Cuervo, Elizabeth P. Derryberry, Travis C. Glenn, Michael G. Harvey, Peter A. Hosner, Leo Joseph, Rebecca Kimball, Andrew L. Mack, Colin M. Miskelly, A. Townsend Peterson, Mark B. Robbins, Frederick H. Sheldon, Luís Fábio Silveira, Brian T. Smith, Noor D. White, Robert G. Moyle, Brant C. Faircloth Corresponding authors: Carl H. Oliveros, Email: [email protected] Brant C. Faircloth, Email: [email protected] This PDF file includes: Supplementary text Figs. S1 to S10 Table S1 to S3 References for SI reference citations Other supplementary materials for this manuscript include the following: Supplementary Files S1 to S3 1 www.pnas.org/cgi/doi/10.1073/pnas.1813206116 Supplementary Information Text Extended Materials and Methods Library preparation and sequencing. We extracted and purified DNA from fresh muscle tissue, liver tissue, or toepad clips from 113 vouchered museum specimens (Supplementary File S1) using the Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s protocol. We quantified DNA extracts using a Qubit fluorometer, and we prepared aliquots of DNA extracted from muscle and liver at 10 ng/µL in 60 µL volume for shearing. We sheared each DNA sample to 400–600 bp using a Qsonica Q800R sonicator for 15–45 cycles, with each cycle running for 20 seconds on and 20 seconds off at 25% amplitude. -

A Study on Genetic Differentiation and Phylogenetic Relationships Among East Asian Titmice (Family Paridae) and Relatives
Jpn. J. Ornithol. 48: 205-218, 2000 A Study on Genetic Differentiation and Phylogenetic Relationships among East Asian Titmice (Family Paridae) and Relatives Noriko OHTA1,2,Seiji KUSUHARA1and Ryozo KAKIZAWA2 1 Graduate School of Science and Technology, Niigata University. Ninomachi 8050,Igarashi, Niigata-shi, Niigata 950-2181,Japan 2 Yamashina Institute for Ornithology, Konoyama 115, Abiko-shi, Chiba 270-1145,Japan By means of protein electrophoresis on 23 loci (15 enzymes), genetic variation and differentiation among eight East Asian Titmice (family Paridae, genus Parus), two long-tailed tits (family Aegithalidae, genus Aegithalos), a nuthatch (family Sittidae, Sitta) and Bearded Tit (family Paradoxornithidae, genus Panurus) were analyzed. A genealogic dendrogram is constructed by the UPGMA method, using genetic distances among the species. The results of the present study suggests that the East Asian Parus species investigated can be divided into four groups, i.e., groups consisting of (1) Parus montanus, (2) P. ater, P. venustulus and P. varius, (3) P. major and P. monticolus, and (4) P. spilonotus and P. holsti. P. spilonotus seems to have a close relationship to P. holsti, but distant relationships to P. major and P. monticolus. Aegithalos, Sitta and Panurus are relatively closely related to each other, whereas the Parus species are shown to have a family level genetic distance in the Passeriformes. In this study, we could not get enough samples of some species (espe- cially P. montanus and P. spilonotus). And since we obtained samples from dealers, there are some possibility that samples were collected from only one population of each species. The data of other samples from several populations and that of other species are needed to determine the phylogeny of the East Asian Parus species. -

Systematic Notes on Asian Birds. 49 a Preliminary Review of the Aegithalidae, Remizidae and Paridae1
Systematic notes on Asian birds. 49 A preliminary review of the Aegithalidae, Remizidae and Paridae1 S. Eck † & J. Martens Eck, S. & J. Martens. Systematic notes on Asian birds. 49. A preliminary review of the Aegithalidae, Remi- zidae and Paridae. Zool. Med. Leiden 80-5 (1), 21.xii.2006: 1-63, fi gs. 1-6, Plates I-III.— ISSN 0024-0672. Siegfried Eck †, formerly Staatliche Sammlungen für Naturkunde Dresden, Museum für Tierkunde, Königsbrücker Landstrasse 159, D-01109 Dresden, Germany. Jochen Martens, Institut für Zoologie, Johannes Gutenberg-Universität, D-55099 Mainz, Germany (e-mail: [email protected]). Key words: Paridae; Aegithalidae; Remizidae; species limits; systematics; taxonomy; eastern Palaearctic region; Indo-Malayan region; morphology; bioacoustics; molecular genetics; secondary contacts; hy- bridisation; introgression. Proposed recent taxonomic changes in Paridae, Aegithalidae, and Remizidae are reviewed within the geographic scope of this series and their reliability is discussed in terms of the Biological Species Concept with respect to secondary contacts, hybridization, introgression, bioacoustics, and molecular genetics. Certain previously unpublished data are added to support the taxonomic decisions. Introduction This review benefi ts from the excellent treatment of these three passerine families in the monograph by Harrap (1996) 2, preceded by detailed work by Vaurie (1957a, b) on the Palearctic species set upon which Snow (1967) based his treatment for Peters’s Checklist of the Birds of the World. Only after the publication of Harrap (op. cit.) has molecular ge- netics begun to furnish information on parid systematics and taxonomy. This has not only given insights into population structure and population genetics of various taxa, but also helped understanding of species evolution and species limits.