MCM6 Indicates Adverse Tumor Features and Poor Outcomes and Promotes G1/S Cell Cycle Progression in Neuroblastoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cell Growth-Regulated Expression of Mammalian MCM5 and MCM6 Genes Mediated by the Transcription Factor E2F
Oncogene (1999) 18, 2299 ± 2309 ã 1999 Stockton Press All rights reserved 0950 ± 9232/99 $12.00 http://www.stockton-press.co.uk/onc Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F Kiyoshi Ohtani1, Ritsuko Iwanaga1, Masataka Nakamura*,1, Masa-aki Ikeda2, Norikazu Yabuta3, Hiromichi Tsuruga3 and Hiroshi Nojima3 1Human Gene Sciences Center, Tokyo Medical and Dental University, Tokyo 113-8510, Japan 2Department of Developmental Biology, Graduate School of Dentistry, Tokyo Medical and Dental University, Tokyo 113-8549, Japan; 3Department of Molecular Genetics, Research Institute for Microbial Diseases, Osaka University, Suita 565-0871, Japan Initiation of DNA replication requires the function of family (MCM2-7) that have been identi®ed in yeast, MCM gene products, which participate in ensuring that Xenopus, and human. Mcm proteins seem to regulate DNA replication occurs only once in the cell cycle. the initiation at the replication origin where the loading Expression of all mammalian genes of the MCM family of the proteins onto the origin recognition complex is induced by growth stimulation, unlike yeast, and the (ORC) is regulated by Cdc6 and cyclin-dependent mRNA levels peak at G1/S boundary. In this study, we kinases (Donovan et al., 1997; Tanaka et al., 1997). examined the transcriptional activities of isolated human However, the mechanism(s) by which Mcm proteins MCM gene promoters. Human MCM5 and MCM6 control the initiation of DNA replication remains promoters with mutation in the E2F sites failed in unclear. promoter regulation following serum stimulation and Xenopus Mcm proteins seem to be able to access exogenous E2F expression. -

Bioinformatics-Based Screening of Key Genes for Transformation of Liver
Jiang et al. J Transl Med (2020) 18:40 https://doi.org/10.1186/s12967-020-02229-8 Journal of Translational Medicine RESEARCH Open Access Bioinformatics-based screening of key genes for transformation of liver cirrhosis to hepatocellular carcinoma Chen Hao Jiang1,2, Xin Yuan1,2, Jiang Fen Li1,2, Yu Fang Xie1,2, An Zhi Zhang1,2, Xue Li Wang1,2, Lan Yang1,2, Chun Xia Liu1,2, Wei Hua Liang1,2, Li Juan Pang1,2, Hong Zou1,2, Xiao Bin Cui1,2, Xi Hua Shen1,2, Yan Qi1,2, Jin Fang Jiang1,2, Wen Yi Gu4, Feng Li1,2,3 and Jian Ming Hu1,2* Abstract Background: Hepatocellular carcinoma (HCC) is the most common type of liver tumour, and is closely related to liver cirrhosis. Previous studies have focussed on the pathogenesis of liver cirrhosis developing into HCC, but the molecular mechanism remains unclear. The aims of the present study were to identify key genes related to the transformation of cirrhosis into HCC, and explore the associated molecular mechanisms. Methods: GSE89377, GSE17548, GSE63898 and GSE54236 mRNA microarray datasets from Gene Expression Omni- bus (GEO) were analysed to obtain diferentially expressed genes (DEGs) between HCC and liver cirrhosis tissues, and network analysis of protein–protein interactions (PPIs) was carried out. String and Cytoscape were used to analyse modules and identify hub genes, Kaplan–Meier Plotter and Oncomine databases were used to explore relationships between hub genes and disease occurrence, development and prognosis of HCC, and the molecular mechanism of the main hub gene was probed using Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway analysis. -

Supplementary Table S1. Correlation Between the Mutant P53-Interacting Partners and PTTG3P, PTTG1 and PTTG2, Based on Data from Starbase V3.0 Database
Supplementary Table S1. Correlation between the mutant p53-interacting partners and PTTG3P, PTTG1 and PTTG2, based on data from StarBase v3.0 database. PTTG3P PTTG1 PTTG2 Gene ID Coefficient-R p-value Coefficient-R p-value Coefficient-R p-value NF-YA ENSG00000001167 −0.077 8.59e-2 −0.210 2.09e-6 −0.122 6.23e-3 NF-YB ENSG00000120837 0.176 7.12e-5 0.227 2.82e-7 0.094 3.59e-2 NF-YC ENSG00000066136 0.124 5.45e-3 0.124 5.40e-3 0.051 2.51e-1 Sp1 ENSG00000185591 −0.014 7.50e-1 −0.201 5.82e-6 −0.072 1.07e-1 Ets-1 ENSG00000134954 −0.096 3.14e-2 −0.257 4.83e-9 0.034 4.46e-1 VDR ENSG00000111424 −0.091 4.10e-2 −0.216 1.03e-6 0.014 7.48e-1 SREBP-2 ENSG00000198911 −0.064 1.53e-1 −0.147 9.27e-4 −0.073 1.01e-1 TopBP1 ENSG00000163781 0.067 1.36e-1 0.051 2.57e-1 −0.020 6.57e-1 Pin1 ENSG00000127445 0.250 1.40e-8 0.571 9.56e-45 0.187 2.52e-5 MRE11 ENSG00000020922 0.063 1.56e-1 −0.007 8.81e-1 −0.024 5.93e-1 PML ENSG00000140464 0.072 1.05e-1 0.217 9.36e-7 0.166 1.85e-4 p63 ENSG00000073282 −0.120 7.04e-3 −0.283 1.08e-10 −0.198 7.71e-6 p73 ENSG00000078900 0.104 2.03e-2 0.258 4.67e-9 0.097 3.02e-2 Supplementary Table S2. -

Cell Cycle-Dependent Modification of Pot1 and Its Effects on Telomere
Cell cycle-dependent modification of Pot1 and its effects on telomere function Vitaliy Kuznetsov Thesis submitted towards the degree of Doctor of Philosophy University College of London Department of Biology London, WC1E 6BT The program of research was carried out at Cancer Research U.K. Laboratory of Telomere Biology 44 Lincoln’s Inn Fields, London, U.K. WC2A 3PX Supervisor: Dr Julia Promisel Cooper September 2008 I, Vitaliy Kuznetsov, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. ABSTRACT Telomere functions are tightly controlled throughout the cell cycle to allow telomerase access while suppressing a bona fide DNA damage response (DDR) at linear chromosome ends. However, the mechanisms that link cell cycle progression with telomere functions are largely unknown. Here we show that a key S-phase kinase, DDK (Dbf4-dependent protein kinase), phosphorylates the telomere binding protein Pot1, and that this phosphorylation is crucial for DNA damage checkpoint inactivation, the suppression of homologous recombination (HR) at telomeres, and the prevention of telomere loss. DDK phosphorylates Pot1 in a very conserved region of its most amino-terminal-proximal OB fold, suggesting that this regulation of telomere function may be widely conserved. Mutation of Pot1 phosphorylation sites leads to telomerase independent telomere maintenance through constant HR, as well as a dependence of telomere maintenance proteins involved in checkpoint activation and HR. These results uncover a novel and important link between DDR suppression and telomere maintenance. The failure in Pot1 phosphorylation and DDR inactivation could potentially lead to uncontrolled cell proliferation without a requirement for telomerase by switching cells to HR dependent telomere homeostasis. -

Prognostic Significance of Minichromosome Maintenance Mrna Expression in Human Lung Adenocarcinoma
ONCOLOGY REPORTS 42: 2279-2292, 2019 Prognostic significance of minichromosome maintenance mRNA expression in human lung adenocarcinoma SHU LI1,2, ZHOU JIANG2, YIRUN LI3 and YANG XU1 1Department of Hematology, 2Cancer Institute (Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education), The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310009; 3Department of General Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310000, P.R. China Received February 20, 2019; Accepted August 9, 2019 DOI: 10.3892/or.2019.7330 Abstract. The minichromosome maintenance (MCM) gene protein-like 4 and anaplastic lymphoma kinase, respec- family plays an essential role in DNA replication and cell cycle tively (4-7). More recently, immunotherapy has been developed progression. However, MCM gene expression has not been and increasingly used in patients with lung cancer, including well-studied in lung adenocarcinoma (LUAD). In the present immune checkpoint inhibitors that target programmed cell study, the expression, prognostic value and functions of MCMs death 1 ligand 1 (PD-L1)-expressing tumor cells by blocking in LUAD were investigated using several databases and PD-L1/PD-1 signaling (8,9). Despite recent advances in bioinformatic tools, including Oncomine, GEPIA, cBioPortal, targeted therapy and immunotherapy, the prognosis of LUAD CancerSEA and Kaplan-Meier plotter. It was demonstrated remains poor (10). It has become a research trend to explore that the mRNA expression of MCM2, MCM4 and MCM10 novel molecular biomarkers or therapeutic targets in the era of were significantly increased in patients with LUAD. High precision medicine (11). In fact, databases based on large-scale, mRNA expression of MCM2-5, MCM8 and MCM10 were genome-wide association studies have facilitated the discovery associated with poor overall survival and progression-free of new biomarkers for cancer management (12). -

Differential and Overlapping Effects of Melatonin and Its Metabolites on Keratinocyte Function: Bioinformatics and Metabolic Analyses
antioxidants Article Differential and Overlapping Effects of Melatonin and Its Metabolites on Keratinocyte Function: Bioinformatics and Metabolic Analyses Joanna Stefan 1,2,† , Tae-Kang Kim 1,†, Fiona Schedel 3, Zorica Janjetovic 1, David K. Crossman 4, Kerstin Steinbrink 3, Radomir M. Slominski 1, Jaroslaw Zmijewski 5, Meri K. Tulic 6, Russel J. Reiter 7 , Konrad Kleszczy ´nski 3,‡ and Andrzej T. Slominski 1,8,*,‡ 1 Department of Dermatology, University of Alabama at Birmingham, Birmingham, AL 35294, USA; [email protected] (J.S.); [email protected] (T.-K.K.); [email protected] (Z.J.); [email protected] (R.M.S.) 2 Department of Oncology, Nicolaus Copernicus University Medical College, Romanowskiej Str. 2, 85-796 Bydgoszcz, Poland 3 Department of Dermatology, University of Münster, Von-Esmarch-Str. 58, 48149 Münster, Germany; [email protected] (F.S.); [email protected] (K.S.); [email protected] (K.K.) 4 Department of Genetics, Comprehensive Cancer Center, University of Alabama at Birmingham, Birmingham, AL 35294, USA; [email protected] 5 Department of Medicine, Comprehensive Cancer Center, University of Alabama at Birmingham, Birmingham, AL 35294, USA; [email protected] 6 Team 12, Centre Méditerranéen de Médecine Moléculaire (C3M), Université Côte d’Azur, INSERM U1065, 06200 Nice, France; [email protected] Citation: Stefan, J.; Kim, T.-K.; 7 Department of Cellular and Structural Biology, UT Health Science Center, San Antonio, TX 78229, USA; Schedel, F.; Janjetovic, Z.; Crossman, [email protected] D.K.; Steinbrink, K.; Slominski, R.M.; 8 Pathology and Laboratory Medicine Service, VA Medical Center, Birmingham, AL 35294, USA Zmijewski, J.; Tulic, M.K.; Reiter, R.J.; * Correspondence: [email protected]; Tel.: +1-205-934-5245; Fax: +1-205-996-0302 et al. -

Eukaryotic Replicative Helicase Subunit Interaction with DNA and Its Role in DNA Replication
G C A T T A C G G C A T genes Review Eukaryotic Replicative Helicase Subunit Interaction with DNA and Its Role in DNA Replication Matthew P. Martinez, Amanda L. Wacker, Irina Bruck and Daniel L. Kaplan * Department of Biomedical Sciences, Florida State University College of Medicine, 1115 W. Call St., Tallahassee, FL 32306, USA; [email protected] (M.P.M.); [email protected] (A.L.W.); [email protected] (I.B.) * Correspondence: [email protected]; Tel.: +1-850-645-0237 Academic Editors: Linda Bloom and Jörg Bungert Received: 17 February 2017; Accepted: 31 March 2017; Published: 6 April 2017 Abstract: The replicative helicase unwinds parental double-stranded DNA at a replication fork to provide single-stranded DNA templates for the replicative polymerases. In eukaryotes, the replicative helicase is composed of the Cdc45 protein, the heterohexameric ring-shaped Mcm2-7 complex, and the tetrameric GINS complex (CMG). The CMG proteins bind directly to DNA, as demonstrated by experiments with purified proteins. The mechanism and function of these DNA-protein interactions are presently being investigated, and a number of important discoveries relating to how the helicase proteins interact with DNA have been reported recently. While some of the protein-DNA interactions directly relate to the unwinding function of the enzyme complex, other protein-DNA interactions may be important for minichromosome maintenance (MCM) loading, origin melting or replication stress. This review describes our current understanding of how the eukaryotic replicative helicase subunits interact with DNA structures in vitro, and proposed models for the in vivo functions of replicative helicase-DNA interactions are also described. -

A Free-Living Protist That Lacks Canonical Eukaryotic DNA Replication and Segregation Systems
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.14.435266; this version posted March 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A free-living protist that lacks canonical eukaryotic DNA replication and segregation systems 2 Dayana E. Salas-Leiva1, Eelco C. Tromer2,3, Bruce A. Curtis1, Jon Jerlström-Hultqvist1, Martin 3 Kolisko4, Zhenzhen Yi5, Joan S. Salas-Leiva6, Lucie Gallot-Lavallée1, Geert J. P. L. Kops3, John M. 4 Archibald1, Alastair G. B. Simpson7 and Andrew J. Roger1* 5 1Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB), Department of 6 Biochemistry and Molecular Biology, Dalhousie University, Halifax, NS, Canada, B3H 4R2 2 7 Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom 8 3Oncode Institute, Hubrecht Institute – KNAW (Royal Netherlands Academy of Arts and Sciences) 9 and University Medical Centre Utrecht, Utrecht, The Netherlands 10 4Institute of Parasitology Biology Centre, Czech Acad. Sci, České Budějovice, Czech Republic 11 5Guangzhou Key Laboratory of Subtropical Biodiversity and Biomonitoring, School of Life Science, 12 South China Normal University, Guangzhou 510631, China 13 6CONACyT-Centro de Investigación en Materiales Avanzados, Departamento de medio ambiente y 14 energía, Miguel de Cervantes 120, Complejo Industrial Chihuahua, 31136 Chihuahua, Chih., México 15 7Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB), Department of 16 Biology, Dalhousie University, Halifax, NS, Canada, B3H 4R2 17 *corresponding author: [email protected] 18 D.E.S-L ORCID iD: 0000-0003-2356-3351 19 E.C.T. -

Cdna Library Screening Identifies Protein Interactors Potentially
ORIGINAL RESEARCH published: 12 November 2015 doi: 10.3389/fpls.2015.00985 cDNA Library Screening Identifies Protein Interactors Potentially Involved in Non-Telomeric Roles of Arabidopsis Telomerase Ladislav Dokládal1,2,DavidHonys3, Rajiv Rana3, Lan-Ying Lee4, Stanton B. Gelvin4 and Eva Sýkorová1,2* 1 Mendel Centre for Plant Genomics and Proteomics, Central European Institute of Technology and Faculty of Science, Masaryk University, Brno, Czech Republic, 2 Institute of Biophysics – Academy of Sciences of the Czech Republic v.v.i., Brno, Czech Republic, 3 Institute of Experimental Botany – Academy of Sciences of the Czech Republic v.v.i., Prague, Czech Republic, 4 Department of Biological Sciences, Purdue University, West Lafayette, IN, USA Telomerase-reverse transcriptase (TERT) plays an essential catalytic role in maintaining telomeres. However, in animal systems telomerase plays additional non-telomeric functional roles. We previously screened an Arabidopsis cDNA library for proteins that interact with the C-terminal extension (CTE) TERT domain and identified a nuclear- localized protein that contains an RNA recognition motif (RRM). This RRM-protein forms homodimers in both plants and yeast. Mutation of the gene encoding the RRM-protein Edited by: Inna Lermontova, had no detectable effect on plant growth and development, nor did it affect telomerase Leibniz Institute of Plant Genetics activity or telomere length in vivo, suggesting a non-telomeric role for TERT/RRM- and Crop Plant Research, Germany protein complexes. The gene encoding the RRM-protein is highly expressed in leaf Reviewed by: and reproductive tissues. We further screened an Arabidopsis cDNA library for proteins Biswapriya Biswavas Misra, University of Florida, USA that interact with the RRM-protein and identified five interactors. -
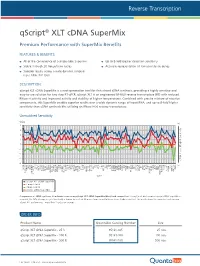
Qscript® XLT Cdna Supermix
-4 -3 -2 -1 -∆Cq Unmatched Sensitivity sensitivity components, thisSuperMixenablessuperiorresultsoverawidedynamicrangeofinputRNA,andupto RNase Hactivityandimprovedstabilityathighertemperatures.Combinedwithprecisemixtureofreaction with reduced reversetranscriptase(RT) isanengineeredM-MLV qScriptXLT easy-to-use solutionfortwostepRT-qPCR. cDNASuperMixisanext-generationtoolforfirst-strandsynthesis,providinghighlysensitiveand qScript XLT DESCRIPTION: qScript XLTperformance-morethan2cyclesonaverage. a control,the Comparison ofcDNAsynthesisfrombraintissueusingqScriptXLTSuperMix (blue)andcompetitors. For more info visit:www.quantabio.com For more Premium PerformancewithSuperMixBenefits qScript FEATURES & BENEFITS: 0 1 2 3 4 Product Name qScript XLT cDNAqScript XLT SuperMix -500 R cDNAqScript XLT SuperMix R -100 cDNAqScript XLT SuperMix R -25 ORDER INFO awidedynamicrangeof across Superior results cycles 20freeze/thaw Stable through All oftheconvenienceasingle-tubeSupermix ACTB input RNA(10 ADAM19 qScript cDNASuperMix Competitor Competitor qScript XLTcDNASuperMi ALDOA APC ARAF ARHGDIA thancDNAsynthesiskitsutilizinganRNaseH(+)reversetranscriptase. ∆ Cq (changeincyclethreshold)isshownforeachof96genesfromapanelbrain tissue(higherisbetter).Theresultsshow ATP6V1G1 B2M BCL2 B A BRCA1 BUB3 8 CASP3 fold) CASP6 CCND1 ® CD40 CD86 cDNA SuperMix XLT CDKN1A x COL1A1 COPS8 CSNK2B CTNNB1 CTSB DFFA DNMT1 DNMT3A DUSP6 EGFR ENSA ERBB2 EZH2 FBXO32 FOXM1 GAPDH GDF11 GTF2I GUSB HK1 HMOX1 HPRT1 HSPA13 HSPA5 HUS1 Quantabio Catalog Number IER2 Gen IER3 IGF2 e IL1R1 IL1RN -

Human Proteins That Interact with RNA/DNA Hybrids
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Resource Human proteins that interact with RNA/DNA hybrids Isabel X. Wang,1,2 Christopher Grunseich,3 Jennifer Fox,1,2 Joshua Burdick,1,2 Zhengwei Zhu,2,4 Niema Ravazian,1 Markus Hafner,5 and Vivian G. Cheung1,2,4 1Howard Hughes Medical Institute, Chevy Chase, Maryland 20815, USA; 2Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, USA; 3Neurogenetics Branch, National Institute of Neurological Disorders and Stroke, NIH, Bethesda, Maryland 20892, USA; 4Department of Pediatrics, University of Michigan, Ann Arbor, Michigan 48109, USA; 5Laboratory of Muscle Stem Cells and Gene Regulation, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Bethesda, Maryland 20892, USA RNA/DNA hybrids form when RNA hybridizes with its template DNA generating a three-stranded structure known as the R-loop. Knowledge of how they form and resolve, as well as their functional roles, is limited. Here, by pull-down assays followed by mass spectrometry, we identified 803 proteins that bind to RNA/DNA hybrids. Because these proteins were identified using in vitro assays, we confirmed that they bind to R-loops in vivo. They include proteins that are involved in a variety of functions, including most steps of RNA processing. The proteins are enriched for K homology (KH) and helicase domains. Among them, more than 300 proteins preferred binding to hybrids than double-stranded DNA. These proteins serve as starting points for mechanistic studies to elucidate what RNA/DNA hybrids regulate and how they are regulated. -

Gene Expression Changes in Cervical Squamous Cancers Following Neoadjuvant Interventional Chemoembolization
Clinica Chimica Acta 493 (2019) 79–86 Contents lists available at ScienceDirect Clinica Chimica Acta journal homepage: www.elsevier.com/locate/cca Gene expression changes in cervical squamous cancers following T neoadjuvant interventional chemoembolization Yonghua Chena,1, Yuanyuan Houa,1, Ying Yanga,1, Meixia Panb, Jing Wanga, Wenshuang Wanga, Ying Zuoa, Jianglin Conga, Xiaojie Wanga, Nan Mua, Chenglin Zhangc, Benjiao Gongc, ⁎ ⁎ Jianqing Houa, , Shaoguang Wanga, , Liping Xud a Department of Obstetrics and Gynecology, the Affiliated Yantai Yuhuangding Hospital of Medical College, Qingdao University, Yantai 264000, Shandong, China b Yantai Yuhuangding Hospital LaiShan Division of Medical College, Qingdao University, China c Central Laboratory, the Affiliated Yantai Yuhuangding Hospital of Medical College, Qingdao University, Yantai 264000, Shandong, China d Medical College, Qingdao University, Qingdao 266021, Shandong, China ARTICLE INFO ABSTRACT Keywords: Background: The efficacy of therapy for cervical cancer is related to the alteration of multiple molecular events Cervical cancer and signaling networks during treatment. The aim of this study was to evaluate gene expression alterations in Chemoembolization advanced cervical cancers before- and after-trans-uterine arterial chemoembolization- (TUACE). Microarray Methods: Gene expression patterns in three squamous cell cervical cancers before- and after-TUACE were de- AKAP12 termined using microarray technique. Changes in AKAP12 and CA9 genes following TUACE were validated by CA9 quantitative real-time PCR. Results: Unsupervised cluster analysis revealed that the after-TUACE samples clustered together, which were separated from the before-TUACE samples. Using a 2-fold threshold, we identified 1131 differentially expressed genes that clearly discriminate after-TUACE tumors from before-TUACE tumors, including 209 up-regulated genes and 922 down-regulated genes.