Phylogenetic Analysis of Sigmodontine Rodents (Muroidea), with Special Reference to the Akodont Genus Deltamys
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ectoparasites of Wild Rodents from Parque Estadual Da Cantareira (Pedra Grande Nuclei), São Paulo, Brazil
ECTOPARASITES OF WILD RODENTS FROM PARQUE ESTADUAL DA CANTAREIRA (PEDRA GRANDE NUCLEI), SÃO PAULO, BRAZIL FERNANDA A. NIERI-BASTOS1 DARCI M. BARROS-BATTESTI1 PEDRO M. LINARDI2 MARCOS AMAKU3 ARLEI MARCILI4 SANDRA E. FAVORITO5; RICARDO PINTO-DA-ROCHA6 ABSTRACT:- NERI-BASTOS, F.A.; BARROS-BATTESTI, D.M.; LINARDI, P.M.; AMAKU, M.; MARCILI, A.; FAVORITO, S.E.; PINTO-DA-ROCHA, R. Ectoparasites of wild rodents from Parque Estadual da Cantareira (Pedra Grande Nuclei), São Paulo, Brazil. [Ectoparasitos de roedores silvestres do Parque Estadual da Cantareira (Núcleo Pedra Grande), São Paulo, Brasil.] Revista Brasileira de Parasitologia Veterinária, v. 13, n. 1, p. 29-35, 2004. Laboratório de Parasitologia, Instituto Butantan, Av. Vital Brasil 1500, São Paulo, SP 05503-900, Brazil. E-mail: [email protected] Sixteen ectoparasite species were collected from 195 wild rodents, between February 2000 and January 2001, in an Ecological Reserve area of the Parque Estadual da Cantareira, in the municipalities of Caieiras, Mariporã and Guarulhos, State of São Paulo, Brazil. Fifty three percent of the captured rodents were found infested, with the highest prevalences observed for the mites Gigantolaelaps gilmorei and G. oudemansi on Oryzomys russatus; G. wolffsohni, Lalelaps paulistanensis and Mysolaelaps parvispinosus on Oligoryzomys sp. In relation to the fleas, Polygenis (Neopolygenis) atopus presented the highest prevalence, infesting Oryzomys russatus. The highest specificity indices were found for Eubrachylaelaps rotundus/Akodon sp.; G. gilmorei and G. oudemansi/O. russatus; and Laelaps navasi/Juliomys pictipes. When average infestation intensities were related to specificity indices, the relationship was only significant for Brucepattersonius sp. and O. russatus (P<0.05). -

Karyotype and Species Diversity of the Genus Delomys (Rodentia, Cricetidae) in Brazil
Acta Theriologica 37 (1 - 2): 163 - 169, 1992. PL ISSN 0001 -7051 Karyotype and species diversity of the genus Delomys (Rodentia, Cricetidae) in Brazil Nilson I. T. ZANCHIN, Ives J. SBALQUEIRO, Alfredo LANGGUTH, Renato C. BOSSLE, Elizabeth C. CASTRO, Luiz F. B. OLIVEIRA and Margarete S. MATTEVI Zanchin N. I. T., Sbalqueiro I. J., Langguth A., Bossle R. C., Castro E. C., Oliveira L. F. B. and Mattevi M. S. 1992. Karyotype and species diversity of the genus Delomys (Rodentia, Cricetidae) in Brazil. Acta theriol. 37: 163 - 169. Cytogenetic analyses were performed in 39 specimens of Delomys trapped in six localities distributed along the Atlantic forest range of the genus. Only two karyotypic forms were found: 2n = 72, FN = 90 to the north and 2n = 82, FN = 80 to the south, with an overlapping area in Sao Paulo and Paraná states. No hybrids were found and given the large difference in karyotype it is likely that any hybrids produced would be infertile. Based on the skin coloration and type localities of the species described it is suggested that the 2n = 72 taxon corresponds to Delomys sublineatus and the 2n = 82 form to D. dorsalis. Departamento de Genética, Universidade Federal do Rio Grande do Sul, C. P. 15053, 91501 Porto Alegre, RS (NITZ, ECC, LFBO, MSM); Departamento de Genética, Universidade Federal do Paraná, Curitiba (IJS, RCB); Museu Nacional, Rio de Janeiro (AL), Brazil. Reprint request should be addressed to MSM Key words: Delomys, karyotypes Introduction The genus Delomys was proposed to include two species of Thomasomys from the southeast coast of Brazil: T. -

Redalyc.Collection Records of Gyldenstolpia Planaltensis (Avila-Pires, 1972) (Rodentia, Cricetidae) Suggest the Local Extinction
Mastozoología Neotropical ISSN: 0327-9383 [email protected] Sociedad Argentina para el Estudio de los Mamíferos Argentina Bezerra, Alexandra M. R. Collection records of Gyldenstolpia planaltensis (Avila-Pires, 1972) (Rodentia, Cricetidae) suggest the local extinction of the species Mastozoología Neotropical, vol. 18, núm. 1, enero-junio, 2011, pp. 119-123 Sociedad Argentina para el Estudio de los Mamíferos Tucumán, Argentina Available in: http://www.redalyc.org/articulo.oa?id=45719986010 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Mastozoología Neotropical, 18(1):119-123, Mendoza, 2011 ISSN 0327-9383 119 ©SAREM, 2011 Versión on-line ISSN 1666-0536 http://www.sarem.org.ar COLLECTION RECORDS OF Gyldenstolpia planaltensis (AVILA-PIRES, 1972) (RODENTIA, CRICETIDAE) SUGGEST THE LOCAL EXTINCTION OF THE SPECIES Alexandra M. R. Bezerra Departamento de Zoologia, Universidade de Brasília, 70910-900, Brasília, DF, Brazil. <abezerra@ fst.com.br> ABSTRACT: The genus Gyldenstolpia was recently described and includes two species, G. fronto and G. planaltensis, both very rare. Gyldenstolpia planaltensis had records in only two localities in the central-western Brazil and is endemic to the Cerrado biome. A new locality is added to G. planaltensis, extending its distribution, and was obtained from a specimen housed in a museum collection. Summarized data on the distribution, ecology and natural history of G. planaltensis denotes that this species can be already extinct or committed to extinction because of habitat conversion and habitat loss. -

A Matter of Weight: Critical Comments on the Basic Data Analysed by Maestri Et Al
DOI: 10.1111/jbi.13098 CORRESPONDENCE A matter of weight: Critical comments on the basic data analysed by Maestri et al. (2016) in Journal of Biogeography, 43, 1192–1202 Abstract Maestri, Luza, et al. (2016), although we believe that an exploration Recently, Maestri, Luza, et al. (2016) assessed the effect of ecology of the quality of the original data informs both. Ultimately, we sub- and phylogeny on body size variation in communities of South mit that the matrix of body size and the phylogeny used by these American Sigmodontinae rodents. Regrettably, a cursory analysis of authors were plagued with major inaccuracies. the data and the phylogeny used to address this question indicates The matrix of body sizes used by Maestri, Luza, et al. (2016, p. that both are plagued with inaccuracies. We urge “big data” users to 1194) was obtained from two secondary or tertiary sources: give due diligence at compiling data in order to avoid developing Rodrıguez, Olalla-Tarraga, and Hawkins (2008) and Bonvicino, Oli- hypotheses based on insufficient or misleading basic information. veira, and D’Andrea (2008). The former study derived cricetid mass data from Smith et al. (2003), an ambitious project focused on the compilation of “body mass information for all mammals on Earth” We are living a great time in evolutionary biology, where the combi- where the basic data were derived from “primary and secondary lit- nation of the increased power of systematics, coupled with the use erature ... Whenever possible, we used an average of male and of ever more inclusive datasets allows—heretofore impossible— female body mass, which was in turn averaged over multiple locali- questions in ecology and evolution to be addressed. -

Check List and Authors Chec List Open Access | Freely Available at Journal of Species Lists and Distribution
ISSN 1809-127X (online edition) © 2010 Check List and Authors Chec List Open Access | Freely available at www.checklist.org.br Journal of species lists and distribution N Mammalia, Rodentia, Cricetidae, Irenomys tarsalis ISTRIBUTIO D gaps (Philippi, 1900): New records for Argentina and filling RAPHIC Gabriel M. Martin G EO G N O E-mail:Consejo [email protected] Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de la Patagonia “San Juan Bosco”, Facultad de Ciencias Naturales Sede Esquel, Laboratorio de Investigaciones en Evolución y Biodiversidad. Sarmiento 849, CP 9200. Esquel, CH, Argentina. OTES N Abstract: Ten new records for the Chilean tree mouse, Irenomys tarsalis, are presented from western Argentina. This at least 125 km in western Chubut Province, where I. tarsalis was previously known for only three records. Additionally, environmentalrepresents a near information 30 % increase at an inecoregional the number and of habitatknown scalelocalities is provided. for the species in this country. Nine of them fill a gap of The Chilean tree mouse Irenomys tarsalis (Philippi, I. tarsalis specimens was made 1900) is a large rodent that inhabits the temperate following Pearson (1995) and by comparison with museum rainforests of southern Chile and adjacent Argentina specimensIdentification (MLP 11.VI.96.10,of MLP 29.IV.99.11), in which (Pearson 1983; 1995; Kelt 1993). Considered one of the combination of the following characters (including the rarest sigmodontines of the Southern Temperate exosomatic, craniomandibular and dental traits) can be Rainforests, the phylogenetic relationship of the species is considered diagnostic (see also Osgood 1943; Mann 1978; still debated and very little is known of its natural history Kelt 1993; Pearson 1995): rat-like in appearance with (Pardiñas et al. -

Intestinal Helminths in Wild Rodents from Native Forest and Exotic Pine Plantations (Pinus Radiata) in Central Chile
animals Communication Intestinal Helminths in Wild Rodents from Native Forest and Exotic Pine Plantations (Pinus radiata) in Central Chile Maira Riquelme 1, Rodrigo Salgado 1, Javier A. Simonetti 2, Carlos Landaeta-Aqueveque 3 , Fernando Fredes 4 and André V. Rubio 1,* 1 Departamento de Ciencias Biológicas Animales, Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, Santa Rosa 11735, La Pintana, Santiago 8820808, Chile; [email protected] (M.R.); [email protected] (R.S.) 2 Departamento de Ciencias Ecológicas, Facultad de Ciencias, Universidad de Chile, Casilla 653, Santiago 7750000, Chile; [email protected] 3 Facultad de Ciencias Veterinarias, Universidad de Concepción, Casilla 537, Chillán 3812120, Chile; [email protected] 4 Departamento de Medicina Preventiva Animal, Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, Santa Rosa 11735, La Pintana, Santiago 8820808, Chile; [email protected] * Correspondence: [email protected]; Tel.: +56-229-780-372 Simple Summary: Land-use changes are one of the most important drivers of zoonotic disease risk in humans, including helminths of wildlife origin. In this paper, we investigated the presence and prevalence of intestinal helminths in wild rodents, comparing this parasitism between a native forest and exotic Monterey pine plantations (adult and young plantations) in central Chile. By analyzing 1091 fecal samples of a variety of rodent species sampled over two years, we recorded several helminth Citation: Riquelme, M.; Salgado, R.; families and genera, some of them potentially zoonotic. We did not find differences in the prevalence of Simonetti, J.A.; Landaeta-Aqueveque, helminths between habitat types, but other factors (rodent species and season of the year) were relevant C.; Fredes, F.; Rubio, A.V. -

Advances in Cytogenetics of Brazilian Rodents: Cytotaxonomy, Chromosome Evolution and New Karyotypic Data
COMPARATIVE A peer-reviewed open-access journal CompCytogenAdvances 11(4): 833–892 in cytogenetics (2017) of Brazilian rodents: cytotaxonomy, chromosome evolution... 833 doi: 10.3897/CompCytogen.v11i4.19925 RESEARCH ARTICLE Cytogenetics http://compcytogen.pensoft.net International Journal of Plant & Animal Cytogenetics, Karyosystematics, and Molecular Systematics Advances in cytogenetics of Brazilian rodents: cytotaxonomy, chromosome evolution and new karyotypic data Camilla Bruno Di-Nizo1, Karina Rodrigues da Silva Banci1, Yukie Sato-Kuwabara2, Maria José de J. Silva1 1 Laboratório de Ecologia e Evolução, Instituto Butantan, Avenida Vital Brazil, 1500, CEP 05503-900, São Paulo, SP, Brazil 2 Departamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo, Rua do Matão 277, CEP 05508-900, São Paulo, SP, Brazil Corresponding author: Maria José de J. Silva ([email protected]) Academic editor: A. Barabanov | Received 1 August 2017 | Accepted 23 October 2017 | Published 21 December 2017 http://zoobank.org/203690A5-3F53-4C78-A64F-C2EB2A34A67C Citation: Di-Nizo CB, Banci KRS, Sato-Kuwabara Y, Silva MJJ (2017) Advances in cytogenetics of Brazilian rodents: cytotaxonomy, chromosome evolution and new karyotypic data. Comparative Cytogenetics 11(4): 833–892. https://doi. org/10.3897/CompCytogen.v11i4.19925 Abstract Rodents constitute one of the most diversified mammalian orders. Due to the morphological similarity in many of the groups, their taxonomy is controversial. Karyotype information proved to be an important tool for distinguishing some species because some of them are species-specific. Additionally, rodents can be an excellent model for chromosome evolution studies since many rearrangements have been described in this group.This work brings a review of cytogenetic data of Brazilian rodents, with information about diploid and fundamental numbers, polymorphisms, and geographical distribution. -

The Pattern of Color Change in Small Mammal
Sandoval Salinas et al. BMC Res Notes (2018) 11:424 https://doi.org/10.1186/s13104-018-3544-x BMC Research Notes RESEARCH NOTE Open Access The pattern of color change in small mammal museum specimens: is it independent of storage histories given museum‑specifc conditions? María Leonor Sandoval Salinas1,2*, José D. Sandoval1,3, Elisa M. Colombo1,3 and Rubén M. Barquez2 Abstract Objective: Determination of color and evaluating its variation form the basis for a broad range of research questions. For studies on taxonomy, systematics, etc., resorting to mammal specimens in museum collections has a number of advantages over using feld specimens. However, if museum specimens are to be for studying color, they should accurately represent the color of live animals, or we should understand how they difer. Basically, this study addresses this question: How does coat color vary when dealing with specimens of Akodon budini (Budin’s grass mouse, Thomas 1918), stored in one museum collection for diferent periods of time? Results: We measured color values through a spectroradiometer and a difuse illumination cabin and used the refectance values in the form of CIELab tri-stimulus values, considering CIE standard illuminant A. We observed that there is a relationship between specimen storage antiquity and pelage color and it seems that it is general for at least a number of small mammals and this could indicate a universal phenomenon across several mammal species and across several storage conditions. Our results, as others, emphasize the importance of considering storage time, among other circumstances, in research studies using mammal skins and where color is of importance. -

Rodentia: Cricetidae: Sigmodontinae) in São Paulo State, Southeastern Brazil: a Locally Extinct Species?
Volume 55(4):69‑80, 2015 THE PRESENCE OF WILFREDOMYS OENAX (RODENTIA: CRICETIDAE: SIGMODONTINAE) IN SÃO PAULO STATE, SOUTHEASTERN BRAZIL: A LOCALLY EXTINCT SPECIES? MARCUS VINÍCIUS BRANDÃO¹ ABSTRACT The Rufous-nosed Mouse Wilfredomys oenax is a rare Sigmodontinae rodent known from scarce records from northern Uruguay and south and southeastern Brazil. This species is under- represented in scientific collections and is currently classified as threathened, being considered extinct at Curitiba, Paraná, the only confirmed locality of the species at southeastern Brazil. Although specimens from São Paulo were already reported, the presence of this species in this state seems to have passed unnoticed in recent literature. Through detailed morphological ana- lyzes of specimens cited in literature, the present work confirms and discusses the presence of this species in São Paulo state from a specimen collected more than 70 years ago. Recently, by the use of modern sampling methods, other rare Sigmodontinae rodents, such as Abrawayomys ruschii, Phaenomys ferrugineous and Rhagomys rufescens, have been recorded to São Paulo state. However, no specimen of Wilfredomys oenax has been recently reported indicating that this species might be locally extinct. The record mentioned here adds another species to the state of São Paulo mammal diversity and reinforces the urgency of studying Wilfredomys oenax. Key-Words: Atlantic Forest; Scientific collection; Threatened species. INTRODUCTION São Paulo is one the most studied states in Brazil regarding to fauna. Mammal lists from this state have Mammal species lists based on voucher-speci- been elaborated since the late XIX century (Von Iher- mens and literature records are essential for offering ing, 1894; Vieira, 1944a, b, 1946, 1950, 1953; Vivo, groundwork to understand a species distribution and 1998). -
Supporting Files
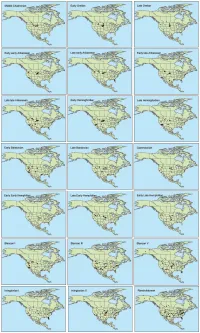
Table S1. Summary of Special Emissions Report Scenarios (SERs) to which we fit climate models for extant mammalian species. Mean Annual Temperature Standard Scenario year (˚C) Deviation Standard Error Present 4.447 15.850 0.057 B1_low 2050s 5.941 15.540 0.056 B1 2050s 6.926 15.420 0.056 A1b 2050s 7.602 15.336 0.056 A2 2050s 8.674 15.163 0.055 A1b 2080s 7.390 15.444 0.056 A2 2080s 9.196 15.198 0.055 A2_top 2080s 11.225 14.721 0.053 Table S2. List of mammalian taxa included and excluded from the species distribution models. -

The Neotropical Region Sensu the Areas of Endemism of Terrestrial Mammals
Australian Systematic Botany, 2017, 30, 470–484 ©CSIRO 2017 doi:10.1071/SB16053_AC Supplementary material The Neotropical region sensu the areas of endemism of terrestrial mammals Elkin Alexi Noguera-UrbanoA,B,C,D and Tania EscalanteB APosgrado en Ciencias Biológicas, Unidad de Posgrado, Edificio A primer piso, Circuito de Posgrados, Ciudad Universitaria, Universidad Nacional Autónoma de México (UNAM), 04510 Mexico City, Mexico. BGrupo de Investigación en Biogeografía de la Conservación, Departamento de Biología Evolutiva, Facultad de Ciencias, Universidad Nacional Autónoma de México (UNAM), 04510 Mexico City, Mexico. CGrupo de Investigación de Ecología Evolutiva, Departamento de Biología, Universidad de Nariño, Ciudadela Universitaria Torobajo, 1175-1176 Nariño, Colombia. DCorresponding author. Email: [email protected] Page 1 of 18 Australian Systematic Botany, 2017, 30, 470–484 ©CSIRO 2017 doi:10.1071/SB16053_AC Table S1. List of taxa processed Number Taxon Number Taxon 1 Abrawayaomys ruschii 55 Akodon montensis 2 Abrocoma 56 Akodon mystax 3 Abrocoma bennettii 57 Akodon neocenus 4 Abrocoma boliviensis 58 Akodon oenos 5 Abrocoma budini 59 Akodon orophilus 6 Abrocoma cinerea 60 Akodon paranaensis 7 Abrocoma famatina 61 Akodon pervalens 8 Abrocoma shistacea 62 Akodon philipmyersi 9 Abrocoma uspallata 63 Akodon reigi 10 Abrocoma vaccarum 64 Akodon sanctipaulensis 11 Abrocomidae 65 Akodon serrensis 12 Abrothrix 66 Akodon siberiae 13 Abrothrix andinus 67 Akodon simulator 14 Abrothrix hershkovitzi 68 Akodon spegazzinii 15 Abrothrix illuteus -

Chromatic Anomalies in Akodontini (Cricetidae: Sigmodontinae) F
Brazilian Journal of Biology https://doi.org/10.1590/1519-6984.214680 ISSN 1519-6984 (Print) Notes and Comments ISSN 1678-4375 (Online) Chromatic anomalies in Akodontini (Cricetidae: Sigmodontinae) F. A. Silvaa,b* , G. Lessab , F. Bertuolc , T. R. O. Freitasd,e and F. M. Quintelaf aPrograma de Pós-graduação em Ecologia e Conservação, Instituto de Biociências, Universidade Federal de Mato Grosso do Sul – UFMS, Cidade Universitária, CEP 79070-900, Campo Grande, MS, Brasil bLaboratório de Mastozoologia, Programa de Pós-graduação em Biologia Animal, Museu de Zoologia João Moojen, Universidade Federal de Viçosa – UFV, Vila Gianetti, Casa 32, Campus Universitário, CEP 36571-000, Viçosa, MG, Brasil cLaboratório de Evolução e Genética Animal, Programa de Pós-graduação em Genética, Conservação e Biologia Evolutiva, Universidade Federal do Amazonas – UFAM, Av. General Rodrigo Otávio Jordão Ramos, 3000, CEP 69077-000, Manaus, AM, Brasil dPrograma de Pós-graduação em Genética e Biologia Molecular, Universidade Federal do Rio Grande do Sul – UFRGS, Av. Bento Gonçalves, 1500, CEP 90040-060, Porto Alegre, RS, Brasil ePrograma de Pós-graduação em Biologia Animal, Departamento de Genética, Universidade Federal do Rio Grande do Sul – UFRGS, Av. Bento Gonçalves, 1500, CEP 90040-060, Porto Alegre, RS, Brasil fLaboratório de Vertebrados, Programa de Pós-graduação em Biologia de Ambientes Aquáticos Continentais, Universidade Federal do Rio Grande – FURG, Av. Itália, CEP 96203-900, Rio Grande, RS, Brasil *e-mail: [email protected] Received: September 27, 2018 – Accepted: February 15, 2019 – Distributed: May 31, 2020 (With 1 figure) The patterns and variability of colors in extant and Álvarez-León, 2014), birds (e. g.