Analysis of Variance---Why It Is More Important Than Ever
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mathematical Statistics
Mathematical Statistics MAS 713 Chapter 1.2 Previous subchapter 1 What is statistics ? 2 The statistical process 3 Population and samples 4 Random sampling Questions? Mathematical Statistics (MAS713) Ariel Neufeld 2 / 70 This subchapter Descriptive Statistics 1.2.1 Introduction 1.2.2 Types of variables 1.2.3 Graphical representations 1.2.4 Descriptive measures Mathematical Statistics (MAS713) Ariel Neufeld 3 / 70 1.2 Descriptive Statistics 1.2.1 Introduction Introduction As we said, statistics is the art of learning from data However, statistical data, obtained from surveys, experiments, or any series of measurements, are often so numerous that they are virtually useless unless they are condensed ; data should be presented in ways that facilitate their interpretation and subsequent analysis Naturally enough, the aspect of statistics which deals with organising, describing and summarising data is called descriptive statistics Mathematical Statistics (MAS713) Ariel Neufeld 4 / 70 1.2 Descriptive Statistics 1.2.2 Types of variables Types of variables Mathematical Statistics (MAS713) Ariel Neufeld 5 / 70 1.2 Descriptive Statistics 1.2.2 Types of variables Types of variables The range of available descriptive tools for a given data set depends on the type of the considered variable There are essentially two types of variables : 1 categorical (or qualitative) variables : take a value that is one of several possible categories (no numerical meaning) Ex. : gender, hair color, field of study, political affiliation, status, ... 2 numerical (or quantitative) -

This History of Modern Mathematical Statistics Retraces Their Development
BOOK REVIEWS GORROOCHURN Prakash, 2016, Classic Topics on the History of Modern Mathematical Statistics: From Laplace to More Recent Times, Hoboken, NJ, John Wiley & Sons, Inc., 754 p. This history of modern mathematical statistics retraces their development from the “Laplacean revolution,” as the author so rightly calls it (though the beginnings are to be found in Bayes’ 1763 essay(1)), through the mid-twentieth century and Fisher’s major contribution. Up to the nineteenth century the book covers the same ground as Stigler’s history of statistics(2), though with notable differences (see infra). It then discusses developments through the first half of the twentieth century: Fisher’s synthesis but also the renewal of Bayesian methods, which implied a return to Laplace. Part I offers an in-depth, chronological account of Laplace’s approach to probability, with all the mathematical detail and deductions he drew from it. It begins with his first innovative articles and concludes with his philosophical synthesis showing that all fields of human knowledge are connected to the theory of probabilities. Here Gorrouchurn raises a problem that Stigler does not, that of induction (pp. 102-113), a notion that gives us a better understanding of probability according to Laplace. The term induction has two meanings, the first put forward by Bacon(3) in 1620, the second by Hume(4) in 1748. Gorroochurn discusses only the second. For Bacon, induction meant discovering the principles of a system by studying its properties through observation and experimentation. For Hume, induction was mere enumeration and could not lead to certainty. Laplace followed Bacon: “The surest method which can guide us in the search for truth, consists in rising by induction from phenomena to laws and from laws to forces”(5). -
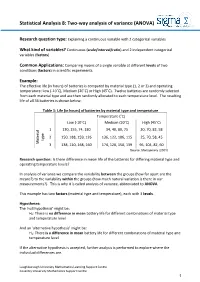
Statistical Analysis 8: Two-Way Analysis of Variance (ANOVA)
Statistical Analysis 8: Two-way analysis of variance (ANOVA) Research question type: Explaining a continuous variable with 2 categorical variables What kind of variables? Continuous (scale/interval/ratio) and 2 independent categorical variables (factors) Common Applications: Comparing means of a single variable at different levels of two conditions (factors) in scientific experiments. Example: The effective life (in hours) of batteries is compared by material type (1, 2 or 3) and operating temperature: Low (-10˚C), Medium (20˚C) or High (45˚C). Twelve batteries are randomly selected from each material type and are then randomly allocated to each temperature level. The resulting life of all 36 batteries is shown below: Table 1: Life (in hours) of batteries by material type and temperature Temperature (˚C) Low (-10˚C) Medium (20˚C) High (45˚C) 1 130, 155, 74, 180 34, 40, 80, 75 20, 70, 82, 58 2 150, 188, 159, 126 136, 122, 106, 115 25, 70, 58, 45 type Material 3 138, 110, 168, 160 174, 120, 150, 139 96, 104, 82, 60 Source: Montgomery (2001) Research question: Is there difference in mean life of the batteries for differing material type and operating temperature levels? In analysis of variance we compare the variability between the groups (how far apart are the means?) to the variability within the groups (how much natural variation is there in our measurements?). This is why it is called analysis of variance, abbreviated to ANOVA. This example has two factors (material type and temperature), each with 3 levels. Hypotheses: The 'null hypothesis' might be: H0: There is no difference in mean battery life for different combinations of material type and temperature level And an 'alternative hypothesis' might be: H1: There is a difference in mean battery life for different combinations of material type and temperature level If the alternative hypothesis is accepted, further analysis is performed to explore where the individual differences are. -
Model Selection for Optimal Prediction in Statistical Machine Learning
Model Selection for Optimal Prediction in Statistical Machine Learning Ernest Fokou´e Introduction science with several ideas from cognitive neuroscience and At the core of all our modern-day advances in artificial in- psychology to inspire the creation, invention, and discov- telligence is the emerging field of statistical machine learn- ery of abstract models that attempt to learn and extract pat- ing (SML). From a very general perspective, SML can be terns from the data. One could think of SML as a field thought of as a field of mathematical sciences that com- of science dedicated to building models endowed with bines mathematics, probability, statistics, and computer the ability to learn from the data in ways similar to the ways humans learn, with the ultimate goal of understand- Ernest Fokou´eis a professor of statistics at Rochester Institute of Technology. His ing and then mastering our complex world well enough email address is [email protected]. to predict its unfolding as accurately as possible. One Communicated by Notices Associate Editor Emilie Purvine. of the earliest applications of statistical machine learning For permission to reprint this article, please contact: centered around the now ubiquitous MNIST benchmark [email protected]. task, which consists of building statistical models (also DOI: https://doi.org/10.1090/noti2014 FEBRUARY 2020 NOTICES OF THE AMERICAN MATHEMATICAL SOCIETY 155 known as learning machines) that automatically learn and Theoretical Foundations accurately recognize handwritten digits from the United It is typical in statistical machine learning that a given States Postal Service (USPS). A typical deployment of an problem will be solved in a wide variety of different ways. -

Applied Time Series Analysis
Applied Time Series Analysis SS 2018 February 12, 2018 Dr. Marcel Dettling Institute for Data Analysis and Process Design Zurich University of Applied Sciences CH-8401 Winterthur Table of Contents 1 INTRODUCTION 1 1.1 PURPOSE 1 1.2 EXAMPLES 2 1.3 GOALS IN TIME SERIES ANALYSIS 8 2 MATHEMATICAL CONCEPTS 11 2.1 DEFINITION OF A TIME SERIES 11 2.2 STATIONARITY 11 2.3 TESTING STATIONARITY 13 3 TIME SERIES IN R 15 3.1 TIME SERIES CLASSES 15 3.2 DATES AND TIMES IN R 17 3.3 DATA IMPORT 21 4 DESCRIPTIVE ANALYSIS 23 4.1 VISUALIZATION 23 4.2 TRANSFORMATIONS 26 4.3 DECOMPOSITION 29 4.4 AUTOCORRELATION 50 4.5 PARTIAL AUTOCORRELATION 66 5 STATIONARY TIME SERIES MODELS 69 5.1 WHITE NOISE 69 5.2 ESTIMATING THE CONDITIONAL MEAN 70 5.3 AUTOREGRESSIVE MODELS 71 5.4 MOVING AVERAGE MODELS 85 5.5 ARMA(P,Q) MODELS 93 6 SARIMA AND GARCH MODELS 99 6.1 ARIMA MODELS 99 6.2 SARIMA MODELS 105 6.3 ARCH/GARCH MODELS 109 7 TIME SERIES REGRESSION 113 7.1 WHAT IS THE PROBLEM? 113 7.2 FINDING CORRELATED ERRORS 117 7.3 COCHRANE‐ORCUTT METHOD 124 7.4 GENERALIZED LEAST SQUARES 125 7.5 MISSING PREDICTOR VARIABLES 131 8 FORECASTING 137 8.1 STATIONARY TIME SERIES 138 8.2 SERIES WITH TREND AND SEASON 145 8.3 EXPONENTIAL SMOOTHING 152 9 MULTIVARIATE TIME SERIES ANALYSIS 161 9.1 PRACTICAL EXAMPLE 161 9.2 CROSS CORRELATION 165 9.3 PREWHITENING 168 9.4 TRANSFER FUNCTION MODELS 170 10 SPECTRAL ANALYSIS 175 10.1 DECOMPOSING IN THE FREQUENCY DOMAIN 175 10.2 THE SPECTRUM 179 10.3 REAL WORLD EXAMPLE 186 11 STATE SPACE MODELS 187 11.1 STATE SPACE FORMULATION 187 11.2 AR PROCESSES WITH MEASUREMENT NOISE 188 11.3 DYNAMIC LINEAR MODELS 191 ATSA 1 Introduction 1 Introduction 1.1 Purpose Time series data, i.e. -

5. Dummy-Variable Regression and Analysis of Variance
Sociology 740 John Fox Lecture Notes 5. Dummy-Variable Regression and Analysis of Variance Copyright © 2014 by John Fox Dummy-Variable Regression and Analysis of Variance 1 1. Introduction I One of the limitations of multiple-regression analysis is that it accommo- dates only quantitative explanatory variables. I Dummy-variable regressors can be used to incorporate qualitative explanatory variables into a linear model, substantially expanding the range of application of regression analysis. c 2014 by John Fox Sociology 740 ° Dummy-Variable Regression and Analysis of Variance 2 2. Goals: I To show how dummy regessors can be used to represent the categories of a qualitative explanatory variable in a regression model. I To introduce the concept of interaction between explanatory variables, and to show how interactions can be incorporated into a regression model by forming interaction regressors. I To introduce the principle of marginality, which serves as a guide to constructing and testing terms in complex linear models. I To show how incremental I -testsareemployedtotesttermsindummy regression models. I To show how analysis-of-variance models can be handled using dummy variables. c 2014 by John Fox Sociology 740 ° Dummy-Variable Regression and Analysis of Variance 3 3. A Dichotomous Explanatory Variable I The simplest case: one dichotomous and one quantitative explanatory variable. I Assumptions: Relationships are additive — the partial effect of each explanatory • variable is the same regardless of the specific value at which the other explanatory variable is held constant. The other assumptions of the regression model hold. • I The motivation for including a qualitative explanatory variable is the same as for including an additional quantitative explanatory variable: to account more fully for the response variable, by making the errors • smaller; and to avoid a biased assessment of the impact of an explanatory variable, • as a consequence of omitting another explanatory variables that is relatedtoit. -

Analysis of Variance and Analysis of Variance and Design of Experiments of Experiments-I
Analysis of Variance and Design of Experimentseriments--II MODULE ––IVIV LECTURE - 19 EXPERIMENTAL DESIGNS AND THEIR ANALYSIS Dr. Shalabh Department of Mathematics and Statistics Indian Institute of Technology Kanpur 2 Design of experiment means how to design an experiment in the sense that how the observations or measurements should be obtained to answer a qqyuery inavalid, efficient and economical way. The desigggning of experiment and the analysis of obtained data are inseparable. If the experiment is designed properly keeping in mind the question, then the data generated is valid and proper analysis of data provides the valid statistical inferences. If the experiment is not well designed, the validity of the statistical inferences is questionable and may be invalid. It is important to understand first the basic terminologies used in the experimental design. Experimental unit For conducting an experiment, the experimental material is divided into smaller parts and each part is referred to as experimental unit. The experimental unit is randomly assigned to a treatment. The phrase “randomly assigned” is very important in this definition. Experiment A way of getting an answer to a question which the experimenter wants to know. Treatment Different objects or procedures which are to be compared in an experiment are called treatments. Sampling unit The object that is measured in an experiment is called the sampling unit. This may be different from the experimental unit. 3 Factor A factor is a variable defining a categorization. A factor can be fixed or random in nature. • A factor is termed as fixed factor if all the levels of interest are included in the experiment. -

Linear Regression: Goodness of Fit and Model Selection
Linear Regression: Goodness of Fit and Model Selection 1 Goodness of Fit I Goodness of fit measures for linear regression are attempts to understand how well a model fits a given set of data. I Models almost never describe the process that generated a dataset exactly I Models approximate reality I However, even models that approximate reality can be used to draw useful inferences or to prediction future observations I ’All Models are wrong, but some are useful’ - George Box 2 Goodness of Fit I We have seen how to check the modelling assumptions of linear regression: I checking the linearity assumption I checking for outliers I checking the normality assumption I checking the distribution of the residuals does not depend on the predictors I These are essential qualitative checks of goodness of fit 3 Sample Size I When making visual checks of data for goodness of fit is important to consider sample size I From a multiple regression model with 2 predictors: I On the left is a histogram of the residuals I On the right is residual vs predictor plot for each of the two predictors 4 Sample Size I The histogram doesn’t look normal but there are only 20 datapoint I We should not expect a better visual fit I Inferences from the linear model should be valid 5 Outliers I Often (particularly when a large dataset is large): I the majority of the residuals will satisfy the model checking assumption I a small number of residuals will violate the normality assumption: they will be very big or very small I Outliers are often generated by a process distinct from those which we are primarily interested in. -

Mathematical Statistics the Sample Distribution of the Median
Course Notes for Math 162: Mathematical Statistics The Sample Distribution of the Median Adam Merberg and Steven J. Miller February 15, 2008 Abstract We begin by introducing the concept of order statistics and ¯nding the density of the rth order statistic of a sample. We then consider the special case of the density of the median and provide some examples. We conclude with some appendices that describe some of the techniques and background used. Contents 1 Order Statistics 1 2 The Sample Distribution of the Median 2 3 Examples and Exercises 4 A The Multinomial Distribution 5 B Big-Oh Notation 6 C Proof That With High Probability jX~ ¡ ¹~j is Small 6 D Stirling's Approximation Formula for n! 7 E Review of the exponential function 7 1 Order Statistics Suppose that the random variables X1;X2;:::;Xn constitute a sample of size n from an in¯nite population with continuous density. Often it will be useful to reorder these random variables from smallest to largest. In reordering the variables, we will also rename them so that Y1 is a random variable whose value is the smallest of the Xi, Y2 is the next smallest, and th so on, with Yn the largest of the Xi. Yr is called the r order statistic of the sample. In considering order statistics, it is naturally convenient to know their probability density. We derive an expression for the distribution of the rth order statistic as in [MM]. Theorem 1.1. For a random sample of size n from an in¯nite population having values x and density f(x), the probability th density of the r order statistic Yr is given by ·Z ¸ ·Z ¸ n! yr r¡1 1 n¡r gr(yr) = f(x) dx f(yr) f(x) dx : (1.1) (r ¡ 1)!(n ¡ r)! ¡1 yr Proof. -

Scalable Model Selection for Spatial Additive Mixed Modeling: Application to Crime Analysis
Scalable model selection for spatial additive mixed modeling: application to crime analysis Daisuke Murakami1,2,*, Mami Kajita1, Seiji Kajita1 1Singular Perturbations Co. Ltd., 1-5-6 Risona Kudan Building, Kudanshita, Chiyoda, Tokyo, 102-0074, Japan 2Department of Statistical Data Science, Institute of Statistical Mathematics, 10-3 Midori-cho, Tachikawa, Tokyo, 190-8562, Japan * Corresponding author (Email: [email protected]) Abstract: A rapid growth in spatial open datasets has led to a huge demand for regression approaches accommodating spatial and non-spatial effects in big data. Regression model selection is particularly important to stably estimate flexible regression models. However, conventional methods can be slow for large samples. Hence, we develop a fast and practical model-selection approach for spatial regression models, focusing on the selection of coefficient types that include constant, spatially varying, and non-spatially varying coefficients. A pre-processing approach, which replaces data matrices with small inner products through dimension reduction dramatically accelerates the computation speed of model selection. Numerical experiments show that our approach selects the model accurately and computationally efficiently, highlighting the importance of model selection in the spatial regression context. Then, the present approach is applied to open data to investigate local factors affecting crime in Japan. The results suggest that our approach is useful not only for selecting factors influencing crime risk but also for predicting crime events. This scalable model selection will be key to appropriately specifying flexible and large-scale spatial regression models in the era of big data. The developed model selection approach was implemented in the R package spmoran. Keywords: model selection; spatial regression; crime; fast computation; spatially varying coefficient modeling 1. -

Model Selection Techniques: an Overview
Model Selection Techniques An overview ©ISTOCKPHOTO.COM/GREMLIN Jie Ding, Vahid Tarokh, and Yuhong Yang n the era of big data, analysts usually explore various statis- following different philosophies and exhibiting varying per- tical models or machine-learning methods for observed formances. The purpose of this article is to provide a compre- data to facilitate scientific discoveries or gain predictive hensive overview of them, in terms of their motivation, large power. Whatever data and fitting procedures are employed, sample performance, and applicability. We provide integrated Ia crucial step is to select the most appropriate model or meth- and practically relevant discussions on theoretical properties od from a set of candidates. Model selection is a key ingredi- of state-of-the-art model selection approaches. We also share ent in data analysis for reliable and reproducible statistical our thoughts on some controversial views on the practice of inference or prediction, and thus it is central to scientific stud- model selection. ies in such fields as ecology, economics, engineering, finance, political science, biology, and epidemiology. There has been a Why model selection long history of model selection techniques that arise from Vast developments in hardware storage, precision instrument researches in statistics, information theory, and signal process- manufacturing, economic globalization, and so forth have ing. A considerable number of methods has been proposed, generated huge volumes of data that can be analyzed to extract useful information. Typical statistical inference or machine- learning procedures learn from and make predictions on data Digital Object Identifier 10.1109/MSP.2018.2867638 Date of publication: 13 November 2018 by fitting parametric or nonparametric models (in a broad 16 IEEE SIGNAL PROCESSING MAGAZINE | November 2018 | 1053-5888/18©2018IEEE sense). -

THE ONE-SAMPLE Z TEST
10 THE ONE-SAMPLE z TEST Only the Lonely Difficulty Scale ☺ ☺ ☺ (not too hard—this is the first chapter of this kind, but youdistribute know more than enough to master it) or WHAT YOU WILL LEARN IN THIS CHAPTERpost, • Deciding when the z test for one sample is appropriate to use • Computing the observed z value • Interpreting the z value • Understandingcopy, what the z value means • Understanding what effect size is and how to interpret it not INTRODUCTION TO THE Do ONE-SAMPLE z TEST Lack of sleep can cause all kinds of problems, from grouchiness to fatigue and, in rare cases, even death. So, you can imagine health care professionals’ interest in seeing that their patients get enough sleep. This is especially the case for patients 186 Copyright ©2020 by SAGE Publications, Inc. This work may not be reproduced or distributed in any form or by any means without express written permission of the publisher. Chapter 10 ■ The One-Sample z Test 187 who are ill and have a real need for the healing and rejuvenating qualities that sleep brings. Dr. Joseph Cappelleri and his colleagues looked at the sleep difficul- ties of patients with a particular illness, fibromyalgia, to evaluate the usefulness of the Medical Outcomes Study (MOS) Sleep Scale as a measure of sleep problems. Although other analyses were completed, including one that compared a treat- ment group and a control group with one another, the important analysis (for our discussion) was the comparison of participants’ MOS scores with national MOS norms. Such a comparison between a sample’s mean score (the MOS score for par- ticipants in this study) and a population’s mean score (the norms) necessitates the use of a one-sample z test.