Phenotypic and Genotypic Characterization of Race TKTTF of Puccinia Graminis F
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
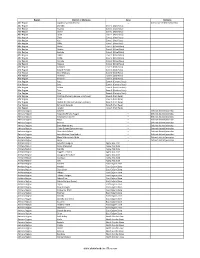
Districts of Ethiopia
Region District or Woredas Zone Remarks Afar Region Argobba Special Woreda -- Independent district/woredas Afar Region Afambo Zone 1 (Awsi Rasu) Afar Region Asayita Zone 1 (Awsi Rasu) Afar Region Chifra Zone 1 (Awsi Rasu) Afar Region Dubti Zone 1 (Awsi Rasu) Afar Region Elidar Zone 1 (Awsi Rasu) Afar Region Kori Zone 1 (Awsi Rasu) Afar Region Mille Zone 1 (Awsi Rasu) Afar Region Abala Zone 2 (Kilbet Rasu) Afar Region Afdera Zone 2 (Kilbet Rasu) Afar Region Berhale Zone 2 (Kilbet Rasu) Afar Region Dallol Zone 2 (Kilbet Rasu) Afar Region Erebti Zone 2 (Kilbet Rasu) Afar Region Koneba Zone 2 (Kilbet Rasu) Afar Region Megale Zone 2 (Kilbet Rasu) Afar Region Amibara Zone 3 (Gabi Rasu) Afar Region Awash Fentale Zone 3 (Gabi Rasu) Afar Region Bure Mudaytu Zone 3 (Gabi Rasu) Afar Region Dulecha Zone 3 (Gabi Rasu) Afar Region Gewane Zone 3 (Gabi Rasu) Afar Region Aura Zone 4 (Fantena Rasu) Afar Region Ewa Zone 4 (Fantena Rasu) Afar Region Gulina Zone 4 (Fantena Rasu) Afar Region Teru Zone 4 (Fantena Rasu) Afar Region Yalo Zone 4 (Fantena Rasu) Afar Region Dalifage (formerly known as Artuma) Zone 5 (Hari Rasu) Afar Region Dewe Zone 5 (Hari Rasu) Afar Region Hadele Ele (formerly known as Fursi) Zone 5 (Hari Rasu) Afar Region Simurobi Gele'alo Zone 5 (Hari Rasu) Afar Region Telalak Zone 5 (Hari Rasu) Amhara Region Achefer -- Defunct district/woredas Amhara Region Angolalla Terana Asagirt -- Defunct district/woredas Amhara Region Artuma Fursina Jile -- Defunct district/woredas Amhara Region Banja -- Defunct district/woredas Amhara Region Belessa -- -

Current Status of Wheat Stem Rust Race Ug99 in South Africa
Current status of wheat stem rust race Ug99 in South Africa Stem rust (black rust) is an important disease of bread wheat in South Africa. It is frequently found in the winter rainfall wheat growing regions of the Western Cape, sometimes reaching epide mic levels and causing significant economic loss. Susceptible cultivars could suffer more than 35% yield loss. The main sign of stem rust is the development of dark-red, elliptical pustules (masses of spores) mainly on the stem, and sometimes on the leaf and head of wheat. Dr Tarekegn Terefe (PTKST), 2SA88+ (TTKSF+) and 2SA42 (PTKSK). ARC-SMALL GRAIN, BETHLEHEM Race TTKSF (virulent on resistance genes Sr9e and Sr38) was first detected in SA in 2000 and LTHOUGH RESISTANT CULTIVARS pro- has dominated the stem rust population over Avide effective stem rust control, the fungus the last 20 years. This race is similar to TTKSP, ex- that causes stem rust frequently acquires new cept for its avirulence on Sr24. The widespread virulence to overcome the resistance in exist- occurrence of TTKSF can be mainly attributed to ing cultivars. The appearance, adaptation and its virulence on several resistance genes and the spread of the stem rust race Ug99 (TTKSK) high- cultivation of susceptible wheat cultivars in the lights the importance of stem rust variability in Western Cape, where stem rust regularly oc- global wheat production. curs. In addition to its broader virulence, TTKSF Ug99 is a highly virulent race which was probably possesses other fitness traits which detected in Uganda, East Africa in 1999. Ug99 may have contributed to its predominance over overcame the resistance gene Sr31 which had the past several years. -

Analysis of Extension Gap Among Improved Bread Wheat Producer's
vv ISSN: 2640-7906 DOI: https://dx.doi.org/10.17352/ojps LIFE SCIENCES GROUP Received: 15 May, 2021 Research Article Accepted: 31 May, 2021 Published: 02 June, 2021 *Corresponding author: Sintayehu Abebe, Ethiopian Analysis of extension gap Institute of Agricultural Research, Kulumsa Agricultural Research Center, Assela, P. O.Box 489, Ethiopia, Tel: +251 9 13 10 42 04; E-mail: among improved bread wheat Keywords: Bread wheat; Extension gap; Clustered- based; Demonstration producer’s farmers found https://www.peertechzpublications.com at Arsi Robe District of Arsi Zone, Oromia Regional State, Ethiopia Sintayehu Abebe* Ethiopian Institute of Agricultural Research, Kulumsa Agricultural Research Center, Assela, P. O.Box 489, Ethiopia Abstract This study reports the analysis of agricultural extension gap among four improved bread wheat varieties at Arsi Robe district, Arsi Zone, along with their management practices under farmers ’condition to enhance farmers’ knowledge and skill on bread wheat production. Four improved bread wheat varieties were used to demonstrate namely Honkolo,DEKA,Hidase(check) and Lemu. Honkolo variety had a 43.8% yield advantage over the standard check (Hidase), DEKA had a 16.4% yield advantage over the standard check (Hidase), and the variety Lemu had a negative -25.8% yield loss or below the standard check (Hidase) and it conclude that both Honkolo and DEKA varieties are the fi rst and second option for the farmers whereas the variety Lemu is not recommended due to low productivity in the study area and others areas having similar agro ecologies. The result of agricultural extension gap analysis showed that Honkolo variety had a -2.7q/ha yield increment or in other words Honkolo variety had achieved 104.3% of the potential at research fi eld and had a 4.3% yield advantage than the research fi eld. -

Protecting Land Tenure Security of Women in Ethiopia: Evidence from the Land Investment for Transformation Program
PROTECTING LAND TENURE SECURITY OF WOMEN IN ETHIOPIA: EVIDENCE FROM THE LAND INVESTMENT FOR TRANSFORMATION PROGRAM Workwoha Mekonen, Ziade Hailu, John Leckie, and Gladys Savolainen Land Investment for Transformation Programme (LIFT) (DAI Global) This research paper was created with funding and technical support of the Research Consortium on Women’s Land Rights, an initiative of Resource Equity. The Research Consortium on Women’s Land Rights is a community of learning and practice that works to increase the quantity and strengthen the quality of research on interventions to advance women’s land and resource rights. Among other things, the Consortium commissions new research that promotes innovations in practice and addresses gaps in evidence on what works to improve women’s land rights. Learn more about the Research Consortium on Women’s Land Rights by visiting https://consortium.resourceequity.org/ This paper assesses the effectiveness of a specific land tenure intervention to improve the lives of women, by asking new questions of available project data sets. ABSTRACT The purpose of this research is to investigate threats to women’s land rights and explore the effectiveness of land certification interventions using evidence from the Land Investment for Transformation (LIFT) program in Ethiopia. More specifically, the study aims to provide evidence on the extent that LIFT contributed to women’s tenure security. The research used a mixed method approach that integrated quantitative and qualitative data. Quantitative information was analyzed from the profiles of more than seven million parcels to understand how the program had incorporated gender interests into the Second Level Land Certification (SLLC) process. -

Small and Medium Forest Enterprises in Ethiopia
The annual value of small and medium forest enterprises (SMFEs) in Ethiopia amounts to hundreds of millions of dollars – dominated in rough order of value by fuelwood, herbal remedies, wild coffee, honey and beeswax and timber furniture. The majority of these enterprises are informal and remain Small and medium forest largely unregulated and untaxed by any government authority. Nevertheless these enterprises appear to have significant social and economic benefits. The Government of Ethiopia has responded by providing support, particularly through enterprises in Ethiopia the framework of Micro and Small Enterprises. The recent establishment of the Oromia State Forest Enterprises Supervising Agency and new policy declarations about the community’s stated role in forest management are clear indications of the current interest in forest resources and the roles they play in rural livelihoods. Non-governmental organisations have also been experimenting with Participatory Forest Management and offered training to emerging enterprises, particularly those engaged in non-timber forest products. Yet few associations have yet been established to try and access the more lucrative markets beyond the local setting. SMFEs have great potential to reduce poverty in Ethiopia, but in their present unregulated state also represent a threat to the country’s declining forest resources. This report consolidates information about them and suggests a practical way forward for those wishing to support them. IIED Small and Medium Forest Enterprise Series No. 26 ISBN -

Total Affected Population, January-June 2012
Total Affected Population, January-June 2012 Erob Tahtay Adiyabo Mereb LekeAhferom Laelay Adiyabo Gulomekeda Dalul Legend · Adwa Saesie Tsaedaemba Werei Leke Tigray HawzenKoneba Lakes Asgede Tsimbila Naeder Adet Atsbi Wenberta Berahle Kola Temben Tselemti Tselemt Administrative boundary Tanqua Abergele Enderta Addi Arekay Ab Ala Afdera Beyeda Saharti Samre Hintalo Wejirat Erebti Abergele Region Janamora Megale Bidu Sahla Alaje Ziquala Endamehoni Sekota Raya Azebo Teru Zone West Belesa Ofla Yalo Gonder Zuria Dehana Kurri Gaz Gibla Alamata Elidar Bugna Gulina Awra Woreda Lasta (Ayna)Gidan Kobo Ewa Afar Lay GayintMeket Wadla Guba Lafto Dubti Total affected Population Delanta Habru Chifra Guba Amhara Worebabu Simada Adaa'r Mile 0(no requirment) MekdelaTenta Afambo Bati z Sayint u Telalak m Mehal Sayint u Legambo Dewa HarewaDewe 14-8108 G Antsokiya Ayisha Sherkole l Kurmuk u Gishe Rabel Sirba Abay g Artuma Fursi Gewane n Menz Gera Midir 8109-13746 Bure Mudaytu a DeraMimo Weremo Jille Timuga Erer Shinile h Afdem is Ibantu Menz Mama Midir Assosa n Wara JarsoHidabu Abote Kewet e Simurobi Gele'alo Dembel 13747-21231 Degem Tarema Ber B Abuna G/BeretKuyu Dire Dawa Ginde Beret Gerar Jarso Miesso Dire DawaJarsoChinaksen Meta Robi DulechaAmibara Goro Gutu Jida Argoba Special KersaHarar Jijiga 21232-33163 JelduAdda Berga Doba Meta Gursum Aleltu Gursum Sasiga Ifata Mieso TuloDeder Babile Hareshen Chiro Zuria Bedeno Diga Mesela Hareri Kebribeyah Addis Ababa Goba KorichaGemechisMalka Balo GirawaFedis 33164-59937 AncharHabro Midega TolaBabile Daro Lebu -

Current Status of Ectoparasites in Sheep and Management Practices
ary Scien in ce r te & e T V e f c h o n n l Journal of VVeterinaryeterinary Science & Bedada et al., J Veterinar Sci Technolo 2015, S:10 o o a a l l n n o o r r g g u u y y DOI: 10.4172/2157-7579.S10-002 o o J J ISSN: 2157-7579 TTechnologyechnology Research Article Open Access Current Status of Ectoparasites in Sheep and Management Practices against the Problem in Ectoparasites Controlled and Uncontrolled Areas of Arsi Zone in Oromia Region, Ethiopia Hailegebriel Bedada, Getachew Terefe and Yacob Hailu Tolossa* College of Veterinary Medicine and Agriculture, Addis Ababa University, Debre Zeit-34, Ethiopia Abstract A cross sectional study was conducted to determine the prevalence of ectoparasites of sheep in ectoparasites controlled and uncontrolled areas, assess major risk factors and evaluate effects of ectoparasites on livelihood of farmer in ectoparasites controlled and uncontrolled areas of Arsi zone. A total of 969 sheep (646 sheep from controlled and 323 sheep from uncontrolled areas) were examined for ectoparasites. From controlled 371 (57.43%) and from uncontrolled area 285 (88.24%) were found to be infested with ectoparasites. The ectoparasites identified in controlled area were B. ovis 48.9%, Linognathus spp 0.93%, sheep keds 7.4%, 2.32% B. decoloratus, 1.46% A. variegatum, 1.08% A. gemma, 4.59% R. evertsi evertsi, and 0.31% mixed ticks infestation and 12.5% mixed infestation with various ectoparasites. Similarly from uncontrolled area identified B. ovis 81.4%, Linognathus spp 0.9%, 1.79% B. -

Putting Ug99 on the Map: an Update on Current and Future Monitoring*
Putting Ug99 on the map: Introduction Detection of stem rust race TTKSK (Ug99) from Uganda An update on current and in 1998/99 (Pretorius et al. 2000) highlighted not only the extremely high vulnerability of the global wheat crop future monitoring* to stem rust but also a lack of adequate global systems to monitor such a threat. The expert panel convened 1 2 3 4 D.P..Hodson ,.K..Nazari ,.R.F..Park ,.J..Hansen ,.P.. by Dr Borlaug in 2005 in response to Ug99 (CIMMYT 4 5 6 4 7 Lassen ,.J..Arista ,.T..Fetch ,.M..Hovmøller ,.Y.Jin ,.Z.A.. 2005) recognized this deficiency and recommended that Pretorius8.and.K..Sonder5 monitoring should be addressed as a priority. The global wheat community responded to this recommendation with Abstract a Global Cereal Rust Monitoring System (GCRMS) being Detection of stem rust race TTKSK (Ug99) from developed by an international consortium within the frame Uganda in 1998/99 highlighted not only the extremely work of the Borlaug Global Rust Initiative (BGRI) (Hodson et high vulnerability of the global wheat crop to stem al. 2009). rust but also a lack of adequate global systems to Concern over races within the Ug99 lineage is justified monitor such a threat. Progress in the development by the well documented historical capacity for stem rust and expansion of the Global Cereal Rust Monitoring to cause severe devastation in all continents in which System (GCRMS) is described. The current situation wheat is grown (see Dubin and Brennan 2009; Hodson regarding the Ug99 lineage of races is outlined and 2011). -

Special Session: UG 99 Status, Management and Prevention
Special Session: UG 99 Status, Management and Prevention Wafa Khoury, AGP-FAO Keith Cressman, AGP-FAO Amor Yahyaoui, ICWIP-ICARDA FAO, Rome 9 April 2008 Current Importance of Wheat Rusts Region Yellow Rust Leaf Rust Stem Rust Australasia Major Local Minor East Asia Major Local Minor South Asia Major Local Minor West Asia Major Local Minor Central Asia Major Major Local Russia/Ukraine Local Major Minor Middle East Major Local Minor North Africa Major Local Minor Eastern Africa Major Local Major Southern Africa Major Local Local Eastern Europe Local Local Minor Western Europe Major Local Minor North America Major Major Minor Central America Major Local Minor South America Local Major Local Special session: UG 99 Status, Management and Prevention Trans-boundary wheat rust Historical information shows records of wind borne spores moving across continents, although shorter distance movements are more common – often within distinct pathozones (or epidemiological regions?). Virulent strain of wheat stripe rust ( Puccinia striiformis ) referred to as Vir.Yr9, appeared in Kenya 1980 then was recovered in Ethiopia in East Africa in 1986 , Pakistan and Nepal in 1990 and reached South Asia in 1993 Spread of Yellow rust followed prevalent current . Special session: UG 99 Status, Management and Prevention Yellow rust (Vir. Yr9) spread 1986-1998 Heavy yield losses were incurred along the pathway of yellow rust from Egypt to Pakistan Special session: UG 99 Status, Management and Prevention Trans-boundary wheat rust In 1999 a new strain of stem rust (Puccinia graminis fsp tritici ) occurred in Uganda, known as Ug99 (www.globalrust.org ). Ug99 represents a much greater threat than Vir Yr9, Estimated 80% of current global wheat varieties are susceptible New Stem rust race: UG99 Stem rust Ug99 followed the expected Ug99-TTKS migration but moved faster to Yemen Ug99: Global threat Can spread from East Africa to Asia, Europe, Australia, America World wheat tsunami???? Such scary reach-out may increase the wheat price in the speculative global market. -

Oromia Region Administrative Map(As of 27 March 2013)
ETHIOPIA: Oromia Region Administrative Map (as of 27 March 2013) Amhara Gundo Meskel ! Amuru Dera Kelo ! Agemsa BENISHANGUL ! Jangir Ibantu ! ! Filikilik Hidabu GUMUZ Kiremu ! ! Wara AMHARA Haro ! Obera Jarte Gosha Dire ! ! Abote ! Tsiyon Jars!o ! Ejere Limu Ayana ! Kiremu Alibo ! Jardega Hose Tulu Miki Haro ! ! Kokofe Ababo Mana Mendi ! Gebre ! Gida ! Guracha ! ! Degem AFAR ! Gelila SomHbo oro Abay ! ! Sibu Kiltu Kewo Kere ! Biriti Degem DIRE DAWA Ayana ! ! Fiche Benguwa Chomen Dobi Abuna Ali ! K! ara ! Kuyu Debre Tsige ! Toba Guduru Dedu ! Doro ! ! Achane G/Be!ret Minare Debre ! Mendida Shambu Daleti ! Libanos Weberi Abe Chulute! Jemo ! Abichuna Kombolcha West Limu Hor!o ! Meta Yaya Gota Dongoro Kombolcha Ginde Kachisi Lefo ! Muke Turi Melka Chinaksen ! Gne'a ! N!ejo Fincha!-a Kembolcha R!obi ! Adda Gulele Rafu Jarso ! ! ! Wuchale ! Nopa ! Beret Mekoda Muger ! ! Wellega Nejo ! Goro Kulubi ! ! Funyan Debeka Boji Shikute Berga Jida ! Kombolcha Kober Guto Guduru ! !Duber Water Kersa Haro Jarso ! ! Debra ! ! Bira Gudetu ! Bila Seyo Chobi Kembibit Gutu Che!lenko ! ! Welenkombi Gorfo ! ! Begi Jarso Dirmeji Gida Bila Jimma ! Ketket Mulo ! Kersa Maya Bila Gola ! ! ! Sheno ! Kobo Alem Kondole ! ! Bicho ! Deder Gursum Muklemi Hena Sibu ! Chancho Wenoda ! Mieso Doba Kurfa Maya Beg!i Deboko ! Rare Mida ! Goja Shino Inchini Sululta Aleltu Babile Jimma Mulo ! Meta Guliso Golo Sire Hunde! Deder Chele ! Tobi Lalo ! Mekenejo Bitile ! Kegn Aleltu ! Tulo ! Harawacha ! ! ! ! Rob G! obu Genete ! Ifata Jeldu Lafto Girawa ! Gawo Inango ! Sendafa Mieso Hirna -

Norman Borlaug
Norman E. Borlaug 1914–2009 A Biographical Memoir by Ronald L. Phillips ©2013 National Academy of Sciences. Any opinions expressed in this memoir are those of the author and do not necessarily reflect the views of the National Academy of Sciences. NORMAN ERNEST BORLAUG March 25, 1914–September 12, 2009 Elected to the NAS, 1968 He cultivated a dream that could empower farmers; He planted the seeds of hope; “ He watered them with enthusiasm; He gave them sunshine; He inspired with his passion; He harvested confidence in the hearts of African farmers; He never gave up. The above words are those of Yohei Sasakawa, chairman of Japan’s Nippon Foundation, written” in memory of Norman E. Borlaug. The author is the son of Ryoichi Sasakawa, who created the Sasakawa Africa Association that applied Borlaug’s work to Africa—the focus of much of the scientist’s efforts in his later years. The passage By Ronald L. Phillips reflects Borlaug’s lifelong philosophy and his tremendous contributions to humanity. His science of wheat breeding, his training of hundreds of developing-country students, and his ability to influence nations to commit to food production are recognized and appreciated around the world. Borlaug was one of only five people to have received all three of the following awards during their lifetimes: the Nobel Peace Prize (1970); the Presidential Medal of Freedom (1977); and the Congressional Gold Medal (2007), which is the highest award that the U.S. government can bestow on a civilian. (The other four were Mother Teresa, Martin Luther King, Nelson Mandela, and Elie Weisel.) Borlaug was elected to the National Academy of Sciences of the United States in 1968 and in 2002 received its Public Welfare Medal, which recognizes “distinguished contributions in the application of science to the public welfare.” At a White House ceremony in 2006, President George W. -

Quantifying Yield Potential and Yield Gaps of Faba Bean in Ethiopia
Ethiop. J. Agric. Sci. 29(3)105-120 (2019) Wondafrash et al. [105] Quantifying Yield Potential and Yield Gaps of Faba Bean in Ethiopia Wondafrash Mulugeta1, Kindie Tesfaye2, Mezegebu Getnet3, Seid Ahmed4, Amsalu Nebiyu1, and Fasil Mekuanint5 1Jimma University College of Agriculture and Veterinary Medicine (JUCAVM) Jimma, Ethiopia. 2International Wheat and Maize Research Center (CIMMYT), Addis Ababa, Ethiopia 3 International Crop Research Institute for the Semi-Arid Tropics (ICRISAT), Addis Ababa, Ethiopia 4 International Centre for Agricultural Research in Dry Areas (ICARDA), Rabat, Morocco. 5 Ethiopian Institute of Agricultural Research (EIAR) አህፅሮት ኢትዮጵያ ባቄላ በዓለም አቀፍ ደረጃ በዋናነት ከሚመረትበቸው አገሮች አንዶ ናት፡፡ ነገር ግን የባቄላ ምርታማነትን በአገር አቀፍ ደረጃ በአማካይ በሄክታር ከ 2 ቶን አይበልጥም፡፡ ለዚህም ዝቅተኛ የዝራያዎች ምርታማነት ፣ ኋላ ቀር የሰብል ጥበቃና አያያዝ ፣ እንዲሁም የአፈር ለምነት መቀነስ እንደ ምክንያት ይጠቀሳሉ ፡፡ የዚህ ምርምር ጥናት ዓላማ የሰብል ዕድገት ሞዴልን በመጠቀም የባቄላን ከፍተኛ ምርታማነትን ካለምንም ማነቆዎች እና በዝናብ ዕጥረት ሁኔታ ያለውን ምርታማነት በመለየት ከአርሶ አደሩ ምርታማነት ጋር ያለውን ክፍተት ማወቅ ነው ፡፡ ለዚህም በኢትዮጵያ ሁኔታ ተስማሚነቱ የተሞከረ Decision Support System for Agrotechnology Transfer(DSSAT)- CROPGRO-faba bean የተባለውን ሞዴል ተጠቅመናል፡፡ ይህም ከመሥክ በተገኘ መረጃ በንፅፅርና በማረጋገጥ ስሌት የተደገፈ ሲሆን የአርሶ አደሩን ምርታማነት ከተፃፉ መዛግብት ወስደናል፡፡ የጥናቱ ውጤት እንደሚያሳየው የባቄላ ምርታማነት ካለምንም ማነቆዎችና በዝናብ ዕጥረት ሁኔታ የምርታማነት ክፍተት በዋና ባቄላ አምራች ዞኖች ከፍተኛ ነው፡፡ በአሁኑ ወቅት በዋና ባቄላ አምራች ዞኖች የሚገኙ አርሶ አደሮች በዝናብ ዕጥረት ሁኔታ ከሚገኘው ምርታማነት በ40 % ያነሰ ምርት ያገኛሉ፡፡ የምርምር ጥናቱ ግኝት እንደሚያመለክተው ከፍተኛ ዝናብ የሚያገኙ ቦታዎች ከፍተኛ የምርታማነት ክፍተት ያሳያሉ ፡፡በተጨማሪ የምርታማነት ክፍተት ደረጃ ምርታማነት አቅምን ለመለየት በምንጠቀምባቸው ዠርያዎች ዓይነት ይወሰናል፡፡ በአጠቃላይ የጥናቱ ግኝት እንደሚያመለክተው የሰብል አያያዝን ትክክለኛና ወቅቱን የጠበቀ የሰብል ጥበቃ በሥራ ላይ በማዋል የባቄላ ምርታማነት ከ 100-300 % መጨመር ይቻላል፡፡ Abstract Ethiopia is one of the major faba bean growing countries in the world but with a low average national yield (≤ 2 t ha-1) compared to yield levels in other countries.