The Puzzling Conservation and Diversification of Lipid Droplets from Bacteria to Eukaryotes Josselin Lupette, Eric Marechal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Oleaginous Green Alga Lobosphaera (Parietochloris) Incisa and Genetic Complementation of a Mutant Strain, Deficient in Arachidonic Acid Biosynthesis
Development of a Nuclear Transformation System for Oleaginous Green Alga Lobosphaera (Parietochloris) incisa and Genetic Complementation of a Mutant Strain, Deficient in Arachidonic Acid Biosynthesis Boris Zorin1., Omer Grundman1., Inna Khozin-Goldberg1*, Stefan Leu1, Michal Shapira2, Yuval Kaye1, Nicolas Tourasse3, Olivier Vallon3, Sammy Boussiba1 1 Microalgal Biotechnology Laboratory, French Associates Institute for Agriculture and Biotechnology of Drylands, J. Blaustein Institutes for Desert Research, Ben-Gurion University of the Negev, Midreshet Ben-Gurion, Israel, 2 Department of Life Sciences, Ben-Gurion University of the Negev, Beer Sheva, Israel, 3 UMR 7141 CNRS/Universite´ Pierre et Marie Curie, Institut de Biologie Physico-Chimique, Paris, France Abstract Microalgae are considered a promising source for various high value products, such as carotenoids, v-3 and v-6 polyunsaturated fatty acids (PUFA). The unicellular green alga Lobosphaera (Parietochloris) incisa is an outstanding candidate for the efficient phototrophic production of arachidonic acid (AA), an essential v-6 PUFA for infant brain development and a widely used ingredient in the baby formula industry. Although phototrophic production of such algal products has not yet been established, estimated costs are considered to be 2–5 times higher than competing heterotrophic production costs. This alga accumulates unprecedented amounts of AA within triacylglycerols and the molecular pathway of AA biosynthesis in L. incisa has been previously elucidated. Thus, progress in transformation and metabolic engineering of this high value alga could be exploited for increasing the efficient production of AA at competitive prices. We describe here the first successful transformation of L. incisa using the ble gene as a selection marker, under the control of the endogenous RBCS promoter. -

Palindromic Genes in the Linear Mitochondrial Genome of the Nonphotosynthetic Green Alga Polytomella Magna
GBE Palindromic Genes in the Linear Mitochondrial Genome of the Nonphotosynthetic Green Alga Polytomella magna David Roy Smith1,y,JimengHua2,3,y, John M. Archibald2, and Robert W. Lee3,* 1Department of Biology, University of Western Ontario, London, Ontario, Canada 2Department of Biochemistry and Molecular Biology, Canadian Institute for Advanced Research, Integrated Microbial Biodiversity Program, Dalhousie University, Halifax, Nova Scotia, Canada 3Department of Biology, Dalhousie University, Halifax, Nova Scotia, Canada *Corresponding author: E-mail: [email protected]. yThese authors contributed equally to this work. Accepted: August 8, 2013 Data deposition: Sequence data from this article have been deposited in GenBank under the accession KC733827. Abstract Organelle DNA is no stranger to palindromic repeats. But never has a mitochondrial or plastid genome been described in which every coding region is part of a distinct palindromic unit. While sequencing the mitochondrial DNA of the nonphotosynthetic green alga Polytomella magna, we uncovered precisely this type of genic arrangement. The P. magna mitochondrial genome is linear and made up entirely of palindromes, each containing 1–7 unique coding regions. Consequently, every gene in the genome is duplicated and in an inverted orientation relative to its partner. And when these palindromic genes are folded into putative stem-loops, their predicted translational start sites are often positioned in the apex of the loop. Gel electrophoresis results support the linear, 28-kb monomeric conformation of the P. magna mitochondrial genome. Analyses of other Polytomella taxa suggest that palindromic mitochondrial genes were present in the ancestor of the Polytomella lineage and lost or retained to various degrees in extant species. -

EMA Strain Catalogue 3Rd Edition
Microalgae strain catalogue A strain selection guide for microalgae users: cultivation and chemical characteristics for high added-value products Gonzalo M. Figueroa-Torres a, Elisabeth Bermejo-Padilla a. Jon K. Pittman b, Constantinos Theodoropoulos a a Department of Chemical Engineering and Analytical Science, Biochemical and Bioprocess Engineering Group b Department of Earth and Environmental Sciences The University of Manchester, Manchester, UK, M13 9PL 3rd Edition Page | 1 Microalgae strain catalogue - A strain selection guide for microalgae users 3rd edition, University of Manchester, Manchester,UK EnhanceMicroAlgae 2021 The 3rd edition of this catalogue contains information on the cultivation and composition characteristics of 37 microalgae. Each entry includes relevant links to Atlantic Area stakeholders known to have a relevant connection with each of the species listed, be it in the form of culture collections, research expertise, technology developers, or biomass producers. We invite the readers to visit and/or join the EnhanceMicroAlgae Stakeholder database: an easily accessible, visual and open access database that brings together all the European Atlantic Area players working in the microalgae sector. Contributing authors: Dr. Gonzalo M. Figueroa-Torres a, Dr. Elisabeth Bermejo-Padilla a. Dr. Jon K. Pittman b, Prof. Constantinos Theodoropoulos a a Department of Chemical Engineering and Analytical Science, Biochemical and Bioprocess Engineering Group b Department of Earth and Environmental Sciences The University of Manchester, Manchester, UK, M13 9PL This publication is part of the deliverables of the Interreg-funded international project EnhanceMicroAlgae. The authors gratefully acknowledge the European Regional Development Fund (ERDF) Interreg Atlantic Area programme which funded the EnhanceMicroAlgae project: EAPA_338/2016, “High added-value industrial opportunities for microalgae in the Atlantic Area”. -

The Apicoplast: a Review of the Derived Plastid of Apicomplexan Parasites
Curr. Issues Mol. Biol. 7: 57-80. Online journalThe Apicoplastat www.cimb.org 57 The Apicoplast: A Review of the Derived Plastid of Apicomplexan Parasites Ross F. Waller1 and Geoffrey I. McFadden2,* way to apicoplast discovery with studies of extra- chromosomal DNAs recovered from isopycnic density 1Botany, University of British Columbia, 3529-6270 gradient fractionation of total Plasmodium DNA. This University Boulevard, Vancouver, BC, V6T 1Z4, Canada group recovered two DNA forms; one a 6kb tandemly 2Plant Cell Biology Research Centre, Botany, University repeated element that was later identifed as the of Melbourne, 3010, Australia mitochondrial genome, and a second, 35kb circle that was supposed to represent the DNA circles previously observed by microscopists (Wilson et al., 1996b; Wilson Abstract and Williamson, 1997). This molecule was also thought The apicoplast is a plastid organelle, homologous to to be mitochondrial DNA, and early sequence data of chloroplasts of plants, that is found in apicomplexan eubacterial-like rRNA genes supported this organellar parasites such as the causative agents of Malaria conclusion. However, as the sequencing effort continued Plasmodium spp. It occurs throughout the Apicomplexa a new conclusion, that was originally embraced with and is an ancient feature of this group acquired by the some awkwardness (“Have malaria parasites three process of endosymbiosis. Like plant chloroplasts, genomes?”, Wilson et al., 1991), began to emerge. apicoplasts are semi-autonomous with their own genome Gradually, evermore convincing character traits of a and expression machinery. In addition, apicoplasts import plastid genome were uncovered, and strong parallels numerous proteins encoded by nuclear genes. These with plastid genomes from non-photosynthetic plants nuclear genes largely derive from the endosymbiont (Epifagus virginiana) and algae (Astasia longa) became through a process of intracellular gene relocation. -

Lateral Gene Transfer of Anion-Conducting Channelrhodopsins Between Green Algae and Giant Viruses
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.15.042127; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 5 Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses Andrey Rozenberg 1,5, Johannes Oppermann 2,5, Jonas Wietek 2,3, Rodrigo Gaston Fernandez Lahore 2, Ruth-Anne Sandaa 4, Gunnar Bratbak 4, Peter Hegemann 2,6, and Oded 10 Béjà 1,6 1Faculty of Biology, Technion - Israel Institute of Technology, Haifa 32000, Israel. 2Institute for Biology, Experimental Biophysics, Humboldt-Universität zu Berlin, Invalidenstraße 42, Berlin 10115, Germany. 3Present address: Department of Neurobiology, Weizmann 15 Institute of Science, Rehovot 7610001, Israel. 4Department of Biological Sciences, University of Bergen, N-5020 Bergen, Norway. 5These authors contributed equally: Andrey Rozenberg, Johannes Oppermann. 6These authors jointly supervised this work: Peter Hegemann, Oded Béjà. e-mail: [email protected] ; [email protected] 20 ABSTRACT Channelrhodopsins (ChRs) are algal light-gated ion channels widely used as optogenetic tools for manipulating neuronal activity 1,2. Four ChR families are currently known. Green algal 3–5 and cryptophyte 6 cation-conducting ChRs (CCRs), cryptophyte anion-conducting ChRs (ACRs) 7, and the MerMAID ChRs 8. Here we 25 report the discovery of a new family of phylogenetically distinct ChRs encoded by marine giant viruses and acquired from their unicellular green algal prasinophyte hosts. -

Evaluating Amplicon High–Throughput Sequencing Data of Microalgae Living in Melting Snow: Improvements and Limitations
Fottea, Olomouc, 19(2): 115–131, 2019 115 DOI: 10.5507/fot.2019.003 Evaluating amplicon high–throughput sequencing data of microalgae living in melting snow: improvements and limitations Stefanie Lutz1*, Lenka Procházková2*, Liane G. Benning1, 3,4, Linda Nedbalová2,5 & Daniel Remias6 1GFZ German Research Centre for Geosciences, Telegrafenberg, 14473 Potsdam, Germany; *Corresponding author e–mail: [email protected]; current address: Agroscope, Müller-Thurgau-Strasse 29, 8820 Wädenswil, Switzerland 2 Department of Ecology, Faculty of Science, Charles University, Viničná 7, 128 44 Prague 2, Czech Republic; *Corresponding author e–mail: [email protected] 3 School of Earth & Environment, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK 4 Department of Earth Sciences, Free University of Berlin, 12249 Berlin, Germany 5 The Czech Academy of Sciences, Institute of Botany, Dukelská 135, 379 82 Třeboň, Czech Republic 6 University of Applied Sciences Upper Austria, Stelzhamerstr. 23, 4600 Wels, Austria Abstract: Melting snowfields are dominated by closely related green algae. Although microscopy–based classifi- cation are evaluable distinction tools, they can be challenging and may not reveal the diversity. High–throughput sequencing (HTS) allows for a comprehensive community evaluation but has been rarely used in such ecosystems. We found that assigning taxonomy to DNA sequences strongly depends on the quality of the reference databases. Furthermore, for an accurate identification, a combination of manual inspection of automated assignments, and oligotyping of the abundant 18S OTUs and ITS2 secondary structure analyses were needed. The use of one marker can be misleading because of low variability (18S) or the scarcity of references (ITS2). -

DGLA from the Microalga Lobosphaera Incsa P127 Modulates Inflammatory Response, Inhibits Inos Expression and Alleviates NO Secretion in RAW264.7 Murine Macrophages
nutrients Article DGLA from the Microalga Lobosphaera Incsa P127 Modulates Inflammatory Response, Inhibits iNOS Expression and Alleviates NO Secretion in RAW264.7 Murine Macrophages Ekaterina Novichkova 1,2, Katya Chumin 3, Noy Eretz-Kdosha 3, Sammy Boussiba 1, Jacob Gopas 4,5 , Guy Cohen 3,6 and Inna Khozin-Goldberg 1,* 1 Microalgal Biotechnology Laboratory, The French Associates Institute for Agriculture and Biotechnology for Drylands, the Jacob Blaustein Institutes for Desert Research, Ben-Gurion University of the Negev, Midreshet Ben-Gurion 8499000, Israel; [email protected] (E.N.); [email protected] (S.B.) 2 The Albert Katz International School for Desert Studies, The Jacob Blaustein Institutes for Desert Research, Ben-Gurion University of the Negev, Midreshet Ben-Gurion 8499000, Israel 3 The Skin Research Institute, The Dead-Sea and Arava Science Centre, Masada 86910, Israel; [email protected] (K.C.); [email protected] (N.E.-K.); [email protected] (G.C.) 4 Department of Microbiology and Immunology and Genetics, Faculty of Health Sciences, Ben-Gurion University of the Negev, Beer Sheva 8400501, Israel; [email protected] 5 Department of Oncology, Soroka University Medical Center, Beer Sheva 8400501, Israel 6 Eilat Campus, Ben-Gurion University of the Negev, Eilat 8855630, Israel * Correspondence: [email protected]; Tel.: +972-8-656-3478 Received: 26 August 2020; Accepted: 18 September 2020; Published: 22 September 2020 Abstract: Microalgae have been considered as a renewable source of nutritional, cosmetic and pharmaceutical compounds. The ability to produce health-beneficial long-chain polyunsaturated fatty acids (LC-PUFA) is of high interest. LC-PUFA and their metabolic lipid mediators, modulate key inflammatory pathways in numerous models. -

Nitrogen Deprivation-Induced Production of Volatile Organic
fmars-07-00410 June 5, 2020 Time: 19:43 # 1 ORIGINAL RESEARCH published: 09 June 2020 doi: 10.3389/fmars.2020.00410 Nitrogen Deprivation-Induced Production of Volatile Organic Compounds in the Arachidonic-Acid-Accumulating Microalga Lobosphaera incisa Underpins Their Role as ROS Scavengers and Chemical Messengers Edited by: Yuji Hiwatashi, Puja Kumari†, Alon Cna’ani, Shoshana Didi-Cohen, Vered Tzin and Miyagi University, Japan Inna Khozin-Goldberg* Reviewed by: French Associates Institute for Agriculture and Biotechnology of Drylands, Jacob Blaustein Institutes for Desert Research, Susana Puntarulo, Ben-Gurion University of the Negev, Beersheba, Israel University of Buenos Aires, Argentina Tiziano Verri, University of Salento, Italy The green microalga Lobosphaera incisa accumulates long-chain polyunsaturated *Correspondence: arachidonic acid sequestered in triacylglycerols under nitrogen (N)-starvation conditions. Inna Khozin-Goldberg [email protected] Many of L. incisa’s physiological and metabolic responses to N-starvation have †Present address: been previously investigated. However, the temporal dynamics of the volatile organic Puja Kumari, compounds (VOCs) under different N availability and their role in L. incisa stress Faculty of Fisheries Sciences, responses have yet to be elucidated. Here, we investigated the VOC profiles of L. Hokkaido University, Hakodate, Japan incisa to reveal their emission patterns, and proposed their physiological roles under Specialty section: N-starvation. Using gas chromatography-mass spectrometry, 42 and 19 VOCs were This article was submitted to Aquatic Physiology, identified in the algal biomass (AVOCs) and in the medium (MVOCs), respectively, a section of the journal belonging to alkanes, alkenes, benzenoids, esters, fatty alcohols, fatty aldehydes, Frontiers in Marine Science fatty acids (FAs), FA esters, ketones, and terpenoids; most of these are the oxidative Received: 31 March 2020 products of FAs or photosynthetic pigment degradation. -

Biogenesis of Lipid Bodies in Lobosphaera Incisa
Biogenesis of Lipid Bodies in Lobosphaera incisa Dissertation for the award of the degree “Doctor rerum naturalium” of the Georg-August-Universität Göttingen within the doctoral program GGNB Microbiology and Biochemistry of the Georg-August University School of Science (GAUSS) submitted by Heike Siegler from Münster Göttingen 2016 Members of the Thesis Committee Prof. Dr. Ivo Feußner Department for Plant Biochemistry, Albrecht-von-Haller Institute for Plant Sciences, University of Göttingen Prof. Dr. Volker Lipka Department of Plant Cell Biology, Albrecht-von-Haller Institute for Plant Sciences, University of Göttingen Prof. Dr. Thomas Friedl Department of Experimental Phycology and Culture Collection of Algae at the University of Göttingen, Albrecht-von-Haller Institute for Plant Sciences, University of Göttingen Members of the Examination Board Prof. Dr. Ivo Feußner (Referee) Department for Plant Biochemistry, Albrecht-von-Haller Institute for Plant Sciences, University of Göttingen Prof. Dr. Volker Lipka (2nd Referee) Department of Plant Cell Biology, Albrecht-von-Haller Institute for Plant Sciences, University of Göttingen Prof. Dr. Thomas Friedl Department of Experimental Phycology and Culture Collection of Algae at the University of Göttingen, Albrecht-von-Haller Institute for Plant Sciences, University of Göttingen Prof. Dr. Andrea Polle Department of Forest Botany and Tree Physiology, Büsgen Institute, University of Göttingen PD Dr. Thomas Teichmann Department of Plant Cell Biology, Albrecht-von-Haller Institute for Plant Sciences, University of Göttingen Dr. Martin Fulda Department for Plant Biochemistry, Albrecht-von-Haller Institute for Plant Sciences, University of Göttingen Date of oral examination: 30.05.2016 Affidavit I hereby declare that I wrote the present dissertation on my own and with no other sources and aids than quoted. -

Plastid-Targeting Peptides from the Chlorarachniophyte Bigelowiella Natans
J. Eukaryot. Microbiol., 51(5), 2004 pp. 529±535 q 2004 by the Society of Protozoologists Plastid-Targeting Peptides from the Chlorarachniophyte Bigelowiella natans MATTHEW B. ROGERS,a JOHN M. ARCHIBALD,a,1 MATTHEW A. FIELD,a CATHERINE LI,b BORIS STRIEPENb and PATRICK J. KEELINGa aCanadian Institute for Advanced Research, Department of Botany, University of British Columbia, 3529-6270 University Boulevard, Vancouver, BC, V6T 1Z4, Canada, and bCenter for Tropical & Emerging Global Diseases & Department of Cellular Biology, University of Georgia, 724 Biological Sciences Building, Athens, Georgia 30602, USA ABSTRACT. Chlorarachniophytes are marine amoebo¯agellate protists that have acquired their plastid (chloroplast) through secondary endosymbiosis with a green alga. Like other algae, most of the proteins necessary for plastid function are encoded in the nuclear genome of the secondary host. These proteins are targeted to the organelle using a bipartite leader sequence consisting of a signal peptide (allowing entry in to the endomembrane system) and a chloroplast transit peptide (for transport across the chloroplast envelope mem- branes). We have examined the leader sequences from 45 full-length predicted plastid-targeted proteins from the chlorarachniophyte Bigelowiella natans with the goal of understanding important features of these sequences and possible conserved motifs. The chemical characteristics of these sequences were compared with a set of 10 B. natans endomembrane-targeted proteins and 38 cytosolic or nuclear proteins, which show that the signal peptides are similar to those of most other eukaryotes, while the transit peptides differ from those of other algae in some characteristics. Consistent with this, the leader sequence from one B. -
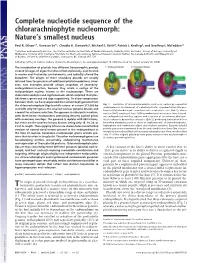
Complete Nucleotide Sequence of the Chlorarachniophyte Nucleomorph: Nature’S Smallest Nucleus
Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature’s smallest nucleus Paul R. Gilson*†, Vanessa Su†‡, Claudio H. Slamovits§, Michael E. Reith¶, Patrick J. Keeling§, and Geoffrey I. McFadden‡ʈ *Infection and Immunity Division, The Walter and Eliza Hall Institute of Medical Research, Parkville 3050, Australia; ‡School of Botany, University of Melbourne, Victoria 3010, Australia; ¶Institute for Marine Biosciences, National Research Council, Halifax, NS, Canada B3H 3Z1; and §Department of Botany, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 Edited by Jeffrey D. Palmer, Indiana University, Bloomington, IN, and approved April 19, 2006 (received for review January 26, 2006) The introduction of plastids into different heterotrophic protists created lineages of algae that diversified explosively, proliferated in marine and freshwater environments, and radically altered the biosphere. The origins of these secondary plastids are usually inferred from the presence of additional plastid membranes. How- ever, two examples provide unique snapshots of secondary- endosymbiosis-in-action, because they retain a vestige of the endosymbiont nucleus known as the nucleomorph. These are chlorarachniophytes and cryptomonads, which acquired their plas- tids from a green and red alga respectively. To allow comparisons between them, we have sequenced the nucleomorph genome from the chlorarachniophyte Bigelowiella natans: at a mere 373,000 bp Fig. 1. Evolution of chlorarachniophytes such as B. natans by sequential endosymbioses. Enslavement of a photosynthetic, cyanobacterium-like pro- and with only 331 genes, the smallest nuclear genome known and karyote (Cb) introduces photosynthesis into a eukaryotic host (Euk 1), whose a model for extreme reduction. The genome is eukaryotic in nature, nucleus (Nu1) acquires at least 1,000 cyanobacterial genes over time. -

Living Organisms Author Their Read-Write Genomes in Evolution
biology Review Living Organisms Author Their Read-Write Genomes in Evolution James A. Shapiro ID Department of Biochemistry and Molecular Biology, University of Chicago GCIS W123B, 979 E. 57th Street, Chicago, IL 60637, USA; [email protected]; Tel.: +1-773-702-1625 Academic Editor: Andrés Moya Received: 23 August 2017; Accepted: 28 November 2017; Published: 6 December 2017 Abstract: Evolutionary variations generating phenotypic adaptations and novel taxa resulted from complex cellular activities altering genome content and expression: (i) Symbiogenetic cell mergers producing the mitochondrion-bearing ancestor of eukaryotes and chloroplast-bearing ancestors of photosynthetic eukaryotes; (ii) interspecific hybridizations and genome doublings generating new species and adaptive radiations of higher plants and animals; and, (iii) interspecific horizontal DNA transfer encoding virtually all of the cellular functions between organisms and their viruses in all domains of life. Consequently, assuming that evolutionary processes occur in isolated genomes of individual species has become an unrealistic abstraction. Adaptive variations also involved natural genetic engineering of mobile DNA elements to rewire regulatory networks. In the most highly evolved organisms, biological complexity scales with “non-coding” DNA content more closely than with protein-coding capacity. Coincidentally, we have learned how so-called “non-coding” RNAs that are rich in repetitive mobile DNA sequences are key regulators of complex phenotypes. Both biotic and abiotic ecological challenges serve as triggers for episodes of elevated genome change. The intersections of cell activities, biosphere interactions, horizontal DNA transfers, and non-random Read-Write genome modifications by natural genetic engineering provide a rich molecular and biological foundation for understanding how ecological disruptions can stimulate productive, often abrupt, evolutionary transformations.