Blueprint Genetics NDUFS2 Single Gene Test
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Proteomic and Metabolomic Analyses of Mitochondrial Complex I-Deficient
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 287, NO. 24, pp. 20652–20663, June 8, 2012 © 2012 by The American Society for Biochemistry and Molecular Biology, Inc. Published in the U.S.A. Proteomic and Metabolomic Analyses of Mitochondrial Complex I-deficient Mouse Model Generated by Spontaneous B2 Short Interspersed Nuclear Element (SINE) Insertion into NADH Dehydrogenase (Ubiquinone) Fe-S Protein 4 (Ndufs4) Gene*□S Received for publication, November 25, 2011, and in revised form, April 5, 2012 Published, JBC Papers in Press, April 25, 2012, DOI 10.1074/jbc.M111.327601 Dillon W. Leong,a1 Jasper C. Komen,b1 Chelsee A. Hewitt,a Estelle Arnaud,c Matthew McKenzie,d Belinda Phipson,e Melanie Bahlo,e,f Adrienne Laskowski,b Sarah A. Kinkel,a,g,h Gayle M. Davey,g William R. Heath,g Anne K. Voss,a,h René P. Zahedi,i James J. Pitt,j Roman Chrast,c Albert Sickmann,i,k Michael T. Ryan,l Gordon K. Smyth,e,f,h b2 a,h,m,n3 David R. Thorburn, and Hamish S. Scott Downloaded from From the aMolecular Medicine Division, gImmunology Division, and eBioinformatics Division, Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria 3052, Australia, the bMurdoch Childrens Research Institute, Royal Children’s Hospital and Department of Paediatrics, University of Melbourne, Parkville, Victoria 3052, Australia, the cDépartement de Génétique Médicale, Université de Lausanne, 1005 Lausanne, Switzerland, the dCentre for Reproduction and Development, Monash Institute of Medical Research, Clayton, Victoria 3168, Australia, the hDepartment of Medical Biology -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Supercomplexes of Prokaryotic Aerobic Respiratory Chains Escherichia Coli and Bacillus Subtilis Supramolecular Assemblies Pedro M.F
Supercomplexes of Prokaryotic Aerobic Respiratory Chains Escherichia coli and Bacillus subtilis supramolecular assemblies Pedro M.F. Sousa Dissertation presented to obtain the Ph.D degree in Biochemistry Instituto de Tecnologia Química e Biológica | Universidade Nova de Lisboa Instituto de Investigação Científica Tropical Oeiras, December, 2013 Supercomplexes of Prokaryotic Aerobic Respiratory Chains Escherichia coli and Bacillus subtilis supramolecular assemblies Pedro M.F. Sousa Dissertation presented to obtain the Ph.D degree in Biochemistry Instituto de Tecnologia Química e Biológica | Universidade Nova de Lisboa Instituto de Investigação Científica Tropical Supervisor: Ana M.P. Melo Co-supervisor: Miguel Teixeira Oeiras, December, 2013 From left to right: Prof. Miguel Teixeira, Dr. Lígia Saraiva, Prof. Carlos Romão, Prof. Thorsten Friedrich, Pedro Sousa, Dr. Margarida Duarte, Prof. João Arrabaça, Dr. Ana Melo. Instituto de Investigação Científica Tropical (IICT) Rua da Junqueira Nº86, 1º 1300-344 Lisboa Portugal Tel. (+351) 213 616 340 Instituto de Tecnologia Química e Biológica (ITQB) Av. da República Estação Agronómica Nacional 2780-157 Oeiras Portugal Tel. (+351) 214 469 323 The molecular cup is now empty. The time has come to replace the purely reductionist “eyes-down” molecular perspective with a new and genuinely holistic “eyes-up” view of the living world, one whose primary focus is on evolution, emergence, and biology’s innate complexity. Carl R.Woese (1928 – 2012) in “A new biology for a new century” Microbiology and Molecular Biology Reviews 68(2), 173 – 186 (2004) Acknowledgements The work summarized in this thesis is the product of several collaborations established during my research grant. It is with great appreciation that I would like to acknowledge the following groups and people: Ana Melo, my supervisor, for her endless support on my work. -

Munkácsy E. & Rea, S. L. the Paradox Of
NIH Public Access Author Manuscript Exp Gerontol. Author manuscript; available in PMC 2015 August 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Exp Gerontol. 2014 August ; 56: 221–233. doi:10.1016/j.exger.2014.03.016. The Paradox of Mitochondrial Dysfunction and Extended Longevity Erin Munkácsy1,2 and Shane L. Rea1,3,* 1Barshop Institute for Longevity and Aging Studies, University of Texas Health Science Center at San Antonio, San Antonio, TX, 78245-3207 USA 2Department of Cell and Structural Biology, University of Texas Health Science Center at San Antonio, San Antonio, TX, 78245-3207 USA 3Department of Physiology, University of Texas Health Science Center at San Antonio, San Antonio, TX, 78245-3207 USA Abstract Mitochondria play numerous, essential roles in the life of eukaryotes. Disruption of mitochondrial function in humans is often pathological or even lethal. Surprisingly, in some organisms mitochondrial dysfunction can result in life extension. This paradox has been studied most extensively in the long-lived Mit mutants of the nematode Caenorhabditis elegans. In this review, we explore the major responses that are activated following mitochondrial dysfunction in these animals and how these responses potentially act to extend their life. We focus our attention on five broad areas of current research – reactive oxygen species signaling, the mitochondrial unfolded protein response, autophagy, metabolic adaptation, and the roles played by various transcription factors. Lastly, we also examine why disruption of complexes I and II differ in their ability to induce the Mit phenotype and extend lifespan. Keywords C. elegans; Mitochondria; Mit Mutant; Lifespan; Aging; Metabolism 1. -

Integrated Proteomic and Mirna Transcriptional Analysis Reveals the Hepatotoxicity Mechanism of PFNA Exposure in Mice
Integrated proteomic and miRNA transcriptional analysis reveals the hepatotoxicity mechanism of PFNA exposure in mice Jianshe Wang, Shengmin Yan, Wei Zhang, Jiayin Dai * Key Laboratory of Animal Ecology and Conservation Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, 100101, P.R. China *Corresponding author: Dai JY; Tel. +86-10-64807185; E-mail: [email protected] Supplementary materials and methods PFNA concentration detection in the serum and liver PFNA in liver and serum samples were quantified using HPLC-MS/MS. Briefly, samples with internal standard were extracted with 5 mL of acetonitrile in 15 mL polypropylene (PP) tubes, and all tubes were placed on a mechanical shaker for 20 min followed by centrifugation at 3,000 × g for 10 min. The top layers, which contained PFNA (analytes and internal standard), were transferred into new PP tubes. The extraction procedure was repeated and a final solution of 10 mL of acetonitrile was combined and concentrated to 0.5 mL under nitrogen gas at 40°C. After the addition of 0.5 mL methanol, the final solution was diluted into 10 mL Milli-Q water for SPE cleanup. All samples were then extracted using an Oasis WAX cartridge (Oasis1 HLB; 150 mg, 6 cc; Waters). The cartridge was pre-equilibrated by the addition of a sequence of 4 mL of 0.1% NH 4OH in methanol, 4 mL methanol, and 4 mL water at a rate of 1 drop per second. Samples (11 mL) were then passed through these cartridges at a rate of 1 drop per second. After loading all samples, cartridges were rinsed with 5 mL Milli-Q water and then washed with 4 mL of 25 mM acetate buffer solution (pH 4). -

Proteome Characteristics of Liver Tissue from Patients with Parenteral
Maitiabola et al. Nutrition & Metabolism (2020) 17:43 https://doi.org/10.1186/s12986-020-00453-z RESEARCH Open Access Proteome characteristics of liver tissue from patients with parenteral nutrition- associated liver disease Gulisudumu Maitiabola1†, Feng Tian1†, Haifeng Sun1†, Li Zhang1, Xuejin Gao1, Bin Xue2* and Xinying Wang1* Abstract Background: Parenteral nutrition (PN)-associated liver disease (PNALD) is a common and life-threatening complication in patients receiving PN. However, its definitive etiology is not yet clear. Therefore, performed proteomic analyses of human liver tissue to explore the same. Methods: Liver tissue was derived and compared across selected patients with (n =3)/without(n =4)PNALDvia isobaric Tag for Relative and Absolute Quantitation (iTRAQ)-based quantitative proteomics. Bioinformatics analysis was performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases to explore the mechanisms of PNALD based on differentially expressed proteins (DEPs). The essential proteins that were differentially expressed between the two groups were explored and verified by western blotting. Results: A total of 112 proteins were found to be differentially expressed, of which 73 were downregulated, and 39 were upregulated in the PNALD group. Bioinformatics analysis showed DEPs to be associated with mitochondrial oxidative phosphorylation (mainly involved in mitochondrial respiratory chain complex I assembly), hepatic glycolipid metabolism (involved primarily in glycogen formation and gluconeogenesis), and oxidative stress (mainly involved in antioxidant change). Conclusion: Overall, our results indicated that mitochondrial energy metabolism impairment, hepatic glycolipid metabolism disorder, and excessive oxidative stress injury might explain the comprehensive mechanism underlying PNALD. Moreover, we have provided multiple potential targets for further exploring the PNALD mechanism. -

A Novel Mutation in NDUFS4 Causes Leigh Syndrome in an Ashkenazi Jewish Family
J Inherit Metab Dis (2008) 31 (Suppl 2):S461–S467 DOI 10.1007/s10545-008-1049-9 SHORT REPORT A novel mutation in NDUFS4 causes Leigh syndrome in an Ashkenazi Jewish family S. L. Anderson & W. K. Chung & J. Frezzo & J. C. Papp & J. Ekstein & S. DiMauro & B. Y. Rubin Received: 15 September 2008 /Submitted in revised form: 13 November 2008 /Accepted: 19 November 2008 / Published online: 26 December 2008 # SSIEM and Springer 2008 Summary Leigh syndrome is a neurodegenerative without consanguinity with three affected children. disorder of infancy or childhood generally due to Linkage to microsatellite markers D5S1969 and mutations in nuclear or mitochondrial genes involved D5S407 led to evaluation of the complex I gene in mitochondrial energy metabolism. We performed NDUFS4, in which we identified a novel homozygous linkage analysis in an Ashkenazi Jewish (AJ) family c.462delA mutation that disrupts the reading frame. The resulting protein lacks a cAMP-dependent protein kinase phosphorylation site required for activation of Communicating editor: John Christodoulou mitochondrial respiratory chain complex I. In a Competing interests: None declared random sample of 5000 healthy AJ individuals, the References to electronic databases: Leigh syndrome: OMIM carrier frequency of the NDUFS4 mutation c.462delA 256000. NDUFS4: OMIM 602694. NDUFS4 mRNA: GenBank was 1 in 1000, suggesting that it should be considered accession # NM_002495. : : in all AJ patients with Leigh syndrome. S. L. Anderson J. Frezzo B. Y. Rubin (*) Department of Biological Sciences, Fordham University, Abbreviations 441 E. Fordham Rd., Bronx, NY 10458, USA AJ Ashkenazi Jewish e-mail: [email protected] BCS1L BCS1-like protein W. -
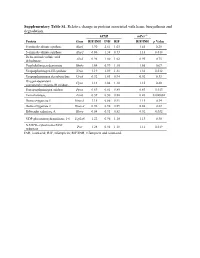
Supplementary Table S1. Relative Change in Proteins Associated with Heme Biosynthesis and Degradation
Supplementary Table S1. Relative change in proteins associated with heme biosynthesis and degradation. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value 5-aminolevulinate synthase Alas1 1.90 2.61 1.05 1.41 0.28 5-aminolevulinate synthase Alas2 0.86 1.38 0.73 1.18 0.018 Delta-aminolevulinic acid Alad 0.96 1.00 1.02 0.95 0.75 dehydratase Porphobilinogen deaminase Hmbs 1.04 0.99 1.10 1.05 0.67 Uroporphyrinogen-III synthase Uros 1.19 1.09 1.31 1.38 0.012 Uroporphyrinogen decarboxylase Urod 0.92 1.03 0.94 0.92 0.33 Oxygen-dependent Cpox 1.13 1.04 1.18 1.15 0.20 coproporphyrinogen-III oxidase, Protoporphyrinogen oxidase Ppox 0.69 0.81 0.85 0.83 0.013 Ferrochelatase, Fech 0.39 0.50 0.88 0.43 0.000002 Heme oxygenase 1 Hmox1 1.15 0.86 0.91 1.11 0.34 Heme oxygenase 2 Hmox2 0.96 0.98 0.89 0.88 0.22 Biliverdin reductase A Blvra 0.84 0.92 0.82 0.92 0.032 UDP-glucuronosyltransferase 1-6 Ugt1a6 1.22 0.96 1.10 1.13 0.30 NADPH--cytochrome P450 Por 1.28 0.92 1.18 1.12 0.019 reductase INH, isoniazid; RIF, rifampicin; RIF/INH, rifampicin and isoniazid. Supplementary Table S2. Relative change in protein nuclear receptors. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value Aryl hydrocarbon receptor Ahr 1.09 0.91 1.00 1.26 0.092 Hepatocyte nuclear factor Hnf1a 0.87 0.97 0.82 0.79 0.027 1-alpha Hepatocyte nuclear factor Hnf4a 0.95 1.05 0.97 1.08 0.20 4-alpha Oxysterols receptor LXR- Nr1h3 0.94 1.16 1.03 1.02 0.42 alpha Bile acid receptor Nr1h4 1.05 1.17 0.98 1.19 0.12 Retinoic acid receptor Rxra 0.88 1.03 0.83 0.95 0.12 RXR-alpha Peroxisome proliferator- -

Reveals of Candidate Active Ingredients in Justicia and Its Anti-Thrombotic Action of Mechanism Based on Network Pharmacology Ap
www.nature.com/scientificreports OPEN Reveals of candidate active ingredients in Justicia and its anti‑thrombotic action of mechanism based on network pharmacology approach and experimental validation Zongchao Hong1,5, Ting Zhang2,5, Ying Zhang1, Zhoutao Xie1, Yi Lu1, Yunfeng Yao1, Yanfang Yang1,3,4*, Hezhen Wu1,3,4* & Bo Liu1,3* Thrombotic diseases seriously threaten human life. Justicia, as a common Chinese medicine, is usually used for anti‑infammatory treatment, and further studies have found that it has an inhibitory efect on platelet aggregation. Therefore, it can be inferred that Justicia can be used as a therapeutic drug for thrombosis. This work aims to reveal the pharmacological mechanism of the anti‑thrombotic efect of Justicia through network pharmacology combined with wet experimental verifcation. During the analysis, 461 compound targets were predicted from various databases and 881 thrombus‑related targets were collected. Then, herb‑compound‑target network and protein–protein interaction network of disease and prediction targets were constructed and cluster analysis was applied to further explore the connection between the targets. In addition, Gene Ontology (GO) and pathway (KEGG) enrichment were used to further determine the association between target proteins and diseases. Finally, the expression of hub target proteins of the core component and the anti‑thrombotic efect of Justicia’s core compounds were verifed by experiments. In conclusion, the core bioactive components, especially justicidin D, can reduce thrombosis by regulating F2, MMP9, CXCL12, MET, RAC1, PDE5A, and ABCB1. The combination of network pharmacology and the experimental research strategies proposed in this paper provides a comprehensive method for systematically exploring the therapeutic mechanism of multi‑component medicine. -

Proteomic Analysis Identifies Distinct Glomerular Extracellular Matrix In
CLINICAL RESEARCH www.jasn.org Proteomic Analysis Identifies Distinct Glomerular Extracellular Matrix in Collapsing Focal Segmental Glomerulosclerosis Michael L. Merchant,1 Michelle T. Barati,1 Dawn J. Caster ,1 Jessica L. Hata,2 Liliane Hobeika,3 Susan Coventry,2 Michael E. Brier,1 Daniel W. Wilkey,1 Ming Li,1 Ilse M. Rood,4 Jeroen K. Deegens,4 Jack F. Wetzels,4 Christopher P. Larsen,5 Jonathan P. Troost,6 Jeffrey B. Hodgin,7 Laura H. Mariani,8 Matthias Kretzler ,8 Jon B. Klein,1,9 and Kenneth R. McLeish1 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background The mechanisms leading to extracellular matrix (ECM) replacement of areas of glomerular capillaries in histologic variants of FSGS are unknown. This study used proteomics to test the hypothesis that glomerular ECM composition in collapsing FSGS (cFSGS) differs from that of other variants. Methods ECM proteins in glomeruli from biopsy specimens of patients with FSGS not otherwise specified (FSGS-NOS) or cFSGS and from normal controls were distinguished and quantified using mass spectrom- etry, verified and localized using immunohistochemistry (IHC) and confocal microscopy, and assessed for gene expression. The analysis also quantified urinary excretion of ECM proteins and peptides. Results Of 58 ECM proteins that differed in abundance between cFSGS and FSGS-NOS, 41 were more abundant in cFSGS and 17 in FSGS-NOS. IHC showed that glomerular tuft staining for cathepsin B, ca- thepsin C, and annexin A3 in cFSGS was significantly greater than in other FSGS variants, in minimal change disease, or in membranous nephropathy. -

Analysis of Human Mutations in the Supernumerary Subunits of Complex I
life Review Analysis of Human Mutations in the Supernumerary Subunits of Complex I Quynh-Chi L. Dang, Duong H. Phan, Abigail N. Johnson, Mukund Pasapuleti, Hind A. Alkhaldi, Fang Zhang and Steven B. Vik * Department of Biological Sciences, Southern Methodist University, Dallas, TX 75287, USA; [email protected] (Q.-C.L.D.); [email protected] (D.H.P.); [email protected] (A.N.J.); [email protected] (M.P.); [email protected] (H.A.A.); [email protected] (F.Z.) * Correspondence: [email protected] Received: 30 October 2020; Accepted: 16 November 2020; Published: 20 November 2020 Abstract: Complex I is the largest member of the electron transport chain in human mitochondria. It comprises 45 subunits and requires at least 15 assembly factors. The subunits can be divided into 14 “core” subunits that carry out oxidation–reduction reactions and proton translocation, as well as 31 additional supernumerary (or accessory) subunits whose functions are less well known. Diminished levels of complex I activity are seen in many mitochondrial disease states. This review seeks to tabulate mutations in the supernumerary subunits of humans that appear to cause disease. Mutations in 20 of the supernumerary subunits have been identified. The mutations were analyzed in light of the tertiary and quaternary structure of human complex I (PDB id = 5xtd). Mutations were found that might disrupt the folding of that subunit or that would weaken binding to another subunit. In some cases, it appeared that no protein was made or, at least, could not be detected. A very common outcome is the lack of assembly of complex I when supernumerary subunits are mutated or missing.