Wild Strawberry) in Sri Lanka Is Restricted to a Small Climatic Envelop Urging Strict Conservation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hydrocotyle Sibthorpioides and H. Batrachium (Araliaceae) New for New York State
Atha, D. 2017. Hydrocotyle sibthorpioides and H. batrachium (Araliaceae) new for New York State. Phytoneuron 2017-56: 1– 6. Published 21 August 2017. ISSN 2153 733X HYDROCOTYLE SIBTHORPIOIDES AND H. BATRACHIUM (ARALIACEAE) NEW FOR NEW YORK STATE DANIEL ATHA Center for Conservation Strategy New York Botanical Garden Bronx, New York 10458 [email protected] ABSTRACT Spontaneous populations of Hydrocotyle sibthorpioides (lawn marsh pennywort) and H. batrachium (Araliaceae) are documented for New York state for the first time. Hydrocotyle batrachium is also new to North America. Hydrocotyle sibthorpioides was first found in 2013 in Queens county; H. batrachium was first found in 2016 in Westchester county. Both species are native to eastern Asia and show potential to be aggressive invaders in southeastern New York, particularly in wetlands. A key to the species of Hydrocotyle in New York State is provided. Prior to this report four species of Hydrocotyle were known from New York state, all of them native and all but one state-listed rarities: H. umbellata – Rare; H. verticillata var. verticillata – Endangered; and H. ranunculoides – Endangered (Young 2010). Among the native New York species, only H. americana occurs in abundance in the state. It is the only species historically reported for New York City and has not been documented for New York City since 1901. The present report documents two additional species for the New York flora, both native to Asia and naturalized in southeastern New York. Fertile herbarium specimens and DNA samples were obtained for all cited specimens and are available for analysis. Hydrocotyle sibthorpioides Lam. Spontaneous populations of Hydrocotyle sibthorpioides in New York were first detected and identified in Queens County by Nick Wagerik in the summer of 2013. -

Chapter 6 ENUMERATION
Chapter 6 ENUMERATION . ENUMERATION The spermatophytic plants with their accepted names as per The Plant List [http://www.theplantlist.org/ ], through proper taxonomic treatments of recorded species and infra-specific taxa, collected from Gorumara National Park has been arranged in compliance with the presently accepted APG-III (Chase & Reveal, 2009) system of classification. Further, for better convenience the presentation of each species in the enumeration the genera and species under the families are arranged in alphabetical order. In case of Gymnosperms, four families with their genera and species also arranged in alphabetical order. The following sequence of enumeration is taken into consideration while enumerating each identified plants. (a) Accepted name, (b) Basionym if any, (c) Synonyms if any, (d) Homonym if any, (e) Vernacular name if any, (f) Description, (g) Flowering and fruiting periods, (h) Specimen cited, (i) Local distribution, and (j) General distribution. Each individual taxon is being treated here with the protologue at first along with the author citation and then referring the available important references for overall and/or adjacent floras and taxonomic treatments. Mentioned below is the list of important books, selected scientific journals, papers, newsletters and periodicals those have been referred during the citation of references. Chronicles of literature of reference: Names of the important books referred: Beng. Pl. : Bengal Plants En. Fl .Pl. Nepal : An Enumeration of the Flowering Plants of Nepal Fasc.Fl.India : Fascicles of Flora of India Fl.Brit.India : The Flora of British India Fl.Bhutan : Flora of Bhutan Fl.E.Him. : Flora of Eastern Himalaya Fl.India : Flora of India Fl Indi. -

State of Wisconsin 2016 Wetland Plant List
5/12/16 State of Wisconsin 2016 Wetland Plant List Lichvar, R.W., D.L. Banks, W.N. Kirchner, and N.C. Melvin. 2016. The National Wetland Plant List: 2016 wetland ratings. Phytoneuron 2016-30: 1-17. Published 28 April 2016. ISSN 2153 733X http://wetland-plants.usace.army.mil/ Trillium cernuum L. (Whip-Poor-Will-Flow er) Photo: Dan Tenaglia List Counts: Wetland MW NCNE Total UPL 91 109 200 FACU 510 534 1044 FAC 272 288 560 FACW 333 317 650 OBL 480 481 961 Rating 1686 1729 1729 User Notes: 1) Plant species not listed are considered UPL for wetland delineation purposes. 2) A few UPL species are listed because they are rated FACU or wetter in at least one Corps Region. 3) Some state boundaries lie within two or more Corps Regions. If a species occurs in one region but not the other, its rating will be shown in one column and the other column will be BLANK. Approved for public release; distribution is unlimited. 1/26 5/12/16 NORTHCENTRAL GREAT LAKES 2016 SUBREGIONAL WETLAND PLANT LIST Scientific Name Authorship Subregion NCNE Common Name Populus tremuloides Michx. NGL = FAC FACU Quaking Aspen Rubus idaeus L. NGL = FAC FACU Common Red Raspberry 2/26 5/12/16 Scientific Name Authorship MW NCNE Common Name Abies balsamea (L.) P. Mill. FACW FAC Balsam Fir Abutilon theophrasti Medik. FACU FACU Velvetleaf Acalypha gracilens Gray FACU FACU Slender Three-Seed-Mercury Acalypha rhomboidea Raf. FACU FACU Common Three-Seed-Mercury Acer negundo L. FAC FAC Ash-Leaf Maple Acer nigrum Michx. -
Taking Green to the Next Level by REBECCA R
A9 SATURDAY, APRIL 23, 2016 Homes&Garden A publication of the Herald-Sun Advertising Department Taking green to the next level BY REBECCA R. NEWSOME, MIRM reen homes, green building, green living... GWho isn’t familiar with “green” today? We know the term as it relates to techniques and materials that produce homes that consume less energy and other resources, facilitate better indoor air quality, and provide a more durable product requiring less maintenance. We’ve been hearing about green for over 10 years now; it’s not a passing fad. In contemplating where we are with green building today, no one comes to mind as more qualified to ask than Michele Myers, President of (healthy and green) M Squared Builders & Designers and both founding and current Chair of Green Home Builders of the Triangle (GHBT). “We’ve been successful throughout the Triangle in embracing green,” says 2016 NGBS Report goes on to Michele. “In fact, North say that the influential drivers Carolina has more green for homeowners surveyed homes certified to National in purchasing a green home Green Building Standards include durability, indoor air (NGBS) than any other state. quality, energy efficiency, water But moving forward, we need efficiency, lower impact to the to do more to educate the environment, and reduced public that green shouldn’t be carbon footprint. an option in home building, “The difference in cost it should be code. A lot of between a Bronze Level builders still think consumers NGBS certified home and a don’t really care about green, non-certified code compliant but nationwide data contradicts home can be as little as 3%,” this… According to the Michele continues. -

Alnus Maritima Grows in Acidic Soils Along Streams That Flow Into the Atlantic Ocean and Delaware Bay
chinquapin The Newsletter of the Southern Appalachian Botanical Society Volume 16, No. 2 Summer 2008 What’s your water footprint? water is much more complex. Perhaps it’s because I live in the suburbs of Atlanta where we are enduring an ‘exceptional’ drought and have been under a total outdoor watering ban “Today, food ties the world together. We make choices every day— for nearly a year that this article from the June 22 Arizona Daily Star By Sarah hamburger or veggie burger, a baked potato or rice, an apple or Garrecht Gassen caught my eye. It seems to apply in the Southeast as well as banana, a glass of beer or wine—that have consequences across the Arizona. I’ve been collecting sink and shower water for other use for a year. globe. Wasting food, or even not finishing the whole pot of coffee, becomes a much bigger deal once we realize how much water goes into ater conservation can begin with your dinner plate. Sure, the things we consume. it looks like merely a hamburger with cheese, a baked potato and a cup of coffee. But look at it in terms of water: “And virtual water should play an important role in deciding what WIt took about 464 gallons of water to produce that quarter pound of industries a community wants to attract and support. The City of beef, 108 gallons to produce a potato, 37 gallons to make 1 ounce of Peoria in Maricopa County has developed a policy that requires the cheese and 37 gallons of water to create your 8-ounce cup of coffee. -

WO 2016/016826 Al 4 February 2016 (04.02.2016) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/016826 Al 4 February 2016 (04.02.2016) P O P C T (51) International Patent Classification: (74) Agent: xyAJ PARK; Level 22, State Insurance Tower, 1 A01H 1/06 (2006.01) C12N 15/61 (2006.01) Willis Street, Wellington (NZ). C12N 15/29 (2006.01) A01H 5/08 (2006.01) (81) Designated States (unless otherwise indicated, for every C12N 15/113 (2010.01) kind of national protection available): AE, AG, AL, AM, (21) International Application Number: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, PCT/IB2015/055743 BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (22) International Filing Date: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, 30 July 2015 (30.07.2015) KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, (25) Filing Language: English MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, (26) Publication Language: English SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, (30) Priority Data: TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. 6282 14 1 August 2014 (01.08.2014) NZ (84) Designated States (unless otherwise indicated, for every (72) Inventors; and kind of regional protection available): ARIPO (BW, GH, (71) Applicants : DARE, Andrew Patrick [NZ/NZ]; 40 Jef GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, ferson Street, Glendowie, Auckland, 1071 (NZ). -
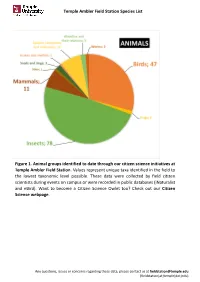
Temple Ambler Field Station Species List Figure 1. Animal Groups Identified to Date Through Our Citizen Science Initiatives at T
Temple Ambler Field Station Species List Figure 1. Animal groups identified to date through our citizen science initiatives at Temple Ambler Field Station. Values represent unique taxa identified in the field to the lowest taxonomic level possible. These data were collected by field citizen scientists during events on campus or were recorded in public databases (iNaturalist and eBird). Want to become a Citizen Science Owlet too? Check out our Citizen Science webpage. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List Figure 2. Plant diversity identified to date in the natural environments and designed gardens of the Temple Ambler Field Station and Ambler Arboretum. These values represent unique taxa identified to the lowest taxonomic level possible. Highlighted are 14 of the 116 flowering plant families present that include 524 taxonomic groups. A full list can be found in our species database. Cultivated specimens in our Greenhouse were not included here. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List database_title Temple Ambler Field Station Species List last_update 22October2020 description This database includes all species identified to their lowest taxonomic level possible in the natural environments and designed gardens on the Temple Ambler campus. These are occurrence records and each taxon is only entered once. This is an occurrence record, not an abundance record. IDs were performed by senior scientists and specialists, as well as citizen scientists visiting campus. -

Quarterly Changes
Plant Names Database: Quarterly changes 30 November 2016 © Landcare Research New Zealand Limited 2016 This copyright work is licensed under the Creative Commons Attribution 4.0 license. Attribution if redistributing to the public without adaptation: "Source: Landcare Research" Attribution if making an adaptation or derivative work: "Sourced from Landcare Research" http://dx.doi.org/doi:10.7931/P19G6B CATALOGUING IN PUBLICATION Plant names database: quarterly changes [electronic resource]. – [Lincoln, Canterbury, New Zealand] : Landcare Research Manaaki Whenua, 2014- . Online resource Quarterly November 2014- ISSN 2382-2341 I.Manaaki Whenua-Landcare Research New Zealand Ltd. II. Allan Herbarium. Citation and Authorship Wilton, A.D.; Schönberger, I.; Gibb, E.S.; Boardman, K.F.; Breitwieser, I.; Cochrane, M.; Dawson, M.I.; de Pauw, B.; Fife, A.J.; Ford, K.A.; Glenny, D.S.; Heenan, P.B.; Korver, M.A.; Novis, P.M.; Redmond, D.N.; Smissen, R.D. Tawiri, K. (2016) Plant Names Database: Quarterly changes. November 2016. Lincoln, Manaaki Whenua Press. This report is generated using an automated system and is therefore authored by the staff at the Allan Herbarium who currently contribute directly to the development and maintenance of the Plant Names Database. Authors are listed alphabetically after the third author. Authors have contributed as follows: Leadership: Wilton, Heenan, Breitwieser Database editors: Wilton, Schönberger, Gibb Taxonomic and nomenclature research and review: Schönberger, Gibb, Wilton, Breitwieser, Dawson, Ford, Fife, Glenny, Heenan, Novis, Redmond, Smissen Information System development: Wilton, De Pauw, Cochrane Technical support: Boardman, Korver, Redmond, Tawiri Disclaimer The Plant Names Database is being updated every working day. We welcome suggestions for improvements, concerns, or any data errors you may find. -

VASCULAR FLORA of the REMNANT BLACKLAND PRAIRIES and ASSOCIATED VEGETATION of GEORGIA by STEPHEN LEE ECHOLS, JR. (Under The
VASCULAR FLORA OF THE REMNANT BLACKLAND PRAIRIES AND ASSOCIATED VEGETATION OF GEORGIA by STEPHEN LEE ECHOLS, JR. (Under the Direction of Wendy B. Zomlefer) ABSTRACT Blackland prairies are a globally imperiled, rare plant community only recently discovered in central Georgia. A floristic inventory was conducted on six remnant blackland prairie sites within Oaky Woods Wildlife Management Area. The 43 ha site complex yielded 354 taxa in 220 genera and 84 families. Four species new to the state of Georgia were documented. Eight rare species, one candidate for federal listing, and one federally endangered species are reported here as new records for the Oaky Woods WMA vicinity. Twelve plant communities are described. A literature review was performed for six states containing blackland prairie within the Gulf and South Atlantic Coastal Plains of the United States. Geology, soils, and vegetation were compared, and cluster analysis was performed using floristic data to assess similarities. INDEX WORDS: Black Belt region, Blackland prairies, cluster analysis, floristics, grassland, Georgia, Oaky Woods Wildlife Management Area, prairie, rare species VASCULAR FLORA OF THE REMNANT BLACKLAND PRAIRIES AND ASSOCIATED VEGETATION OF GEORGIA by STEPHEN LEE ECHOLS, JR. B.S., Appalachian State University, 2002 A Thesis Submitted to the Graduate Faculty of The University of Georgia in Partial Fulfillment of the Requirements for the Degree MASTER OF SCIENCE ATHENS, GEORGIA 2007 © 2007 Stephen Lee Echols, Jr. All Rights Reserved VASCULAR FLORA OF THE REMNANT BLACKLAND PRAIRIES AND ASSOCIATED VEGETATION OF GEORGIA by STEPHEN LEE ECHOLS, JR. Major Professor: Wendy B. Zomlefer Committee: Jim Affolter Rebecca Sharitz Electronic Version Approved: Maureen Grasso Dean of the Graduate School The University of Georgia August 2007 iv DEDICATION I dedicate this thesis to my girlfriend Lisa Keong, who despite my absence, has stood by me with patience and support throughout the final stages of this project. -

Invasive Plant Species in Delaware
NON-NATIVE AND INVASIVE PLANTS IN DELAWARE William A. McAvoy Species Conservation and Research Program Delaware Division of Fish and Wildlife Delaware Dept. of Natural Resources and Environmental Control 4876 Hay Point Landing Rd., Smyrna, DE 19977 [email protected] March 2016 This list consists of 721 species, varieties and subspecies (taxa) of non-native plants, which represents 31% of the Delaware’s flora (2324 total taxa, 1603 taxa are native). Of the 721 non-native taxa listed, 79 are classified as invasive and 188 are watchlist species. Non-native: A species that is not native to North America (north of Mexico). Non-native species are thought to have been introduced by humans, primarily through agricultural or horticultural practices. These species have become established in Delaware and are reproducing as if native (i.e., naturalized). Adventive (highlighted in bold): A species native to North America, but not to Delaware that is now found growing in Delaware outside of its natural range. Adventive species are not considered to be part of Delaware's native flora. These species usually arrive due to the human-caused breakdown of natural barriers to dispersal. Adventive species also include plants that have been introduced, or intentionally planted in Delaware and are now escaping and surviving without cultivation. Invasive: A species that causes environmental harm. Invasive species are very aggressive and out- compete and displace native flora and fauna. Invasive Watchlist: Invasive species that are not yet well established in Delaware but have the potential to become abundant and widely distributed throughout the state. Invasive Watchlist species are often a priority for concern in surrounding states. -
Vascular Flora of the Shoal Creek Preserve Forever Wild Tract, Lauderdale County, Alabama
Holt, B.D., A.S. Peters, C.T. Taylor, and T.W. Barger. 2016. Vascular flora of the Shoal Creek Preserve Forever Wild Tract, Lauderdale County, Alabama. Phytoneuron 2016-6: 1–22. VASCULAR FLORA OF THE SHOAL CREEK PRESERVE FOREVER WILD TRACT, LAUDERDALE COUNTY, ALABAMA 1* 1 1 1 BRIAN D. HOLT , ASHLEY S. PETERS , CHRIS T. TAYLOR , AND T. WAYNE BARGER 1State Lands Division, Natural Heritage Section, Department of Conservation and Natural Resources, 64 North Union Street Montgomery, Alabama 36130 *Correspondence : Brian D. Holt ([email protected]) ABSTRACT The Shoal Creek Preserve Forever Wild Tract (SCPFWT) is a 123 ha property acquired by Alabama’s Forever Wild Land Program on February 26, 2003. The SCPFWT lies 13 km north-northeast of Florence, Alabama, 46 km southeast of Lawrenceburg, Tennessee, and is bordered for a short distance on the east by Shoal Creek. The site is managed by the Alabama Department of Conservation and Natural Resources for habitat conservation, outdoor recreation, and education. An intensive floristic study of this area was conducted from May 2009 through May 2011 and less frequently through June 2015. A total of 519 taxa from 346 genera and 121 families were collected, with 217 taxa representing county records. Asteraceae was the most collected family, with 69 species; Poaceae, Fabaceae, and Cyperaceae were the next largest families with 38, 29, and 17 species, respectively. Carex was the largest genus represented with 11 species. 71 non-native taxa were collected during the surveys. 42 percent of the total collections were county records and 37 exotic taxa collected during this survey were county records. -
Berkov's Full Testimony with Facts About
Dr. Amy Berkov, [email protected] City College of New York, Dept. Biology I don’t understand how the city can even propose an Environmental Impact Statement for a plan that is unprecedented in its destruction: how can anyone predict impact, when there is no precedent? In my area of expertise, terrestrial biodiversity, the DEIS is incomplete, inaccurate and makes unjustified assumptions. 1) The incomplete and inaccurate assessment is based on two four-hour mid-summer walkthroughs, supplemented with external information (not site-specific). The walkthroughs documented approximately 18 bird and insect species (DEIS Appendix F1: "invertebrate species richness was low, as anticipated for a heavily-disturbed, urban system"). According to DEIS 5.6-31, the USFWS iPaC tool gave list of 58 migratory birds that might use the ESCR region. Citizen Scientists have documented >430 species, including 82 bird and 104 insect species. In DEIS Appendix H1, the NYS Natural Heritage Program lists the peregrine falcon as an endangered species known to nest on the Williamsburg bridge. The letter from NYSNHP points out: “For most sites, comprehensive field surveys have not been conducted… further information from on-site surveys or other sources may be required to fully assess impacts on biological resources.” Citizen Scientists have documented nine birds and one bumble bee that are on the NYSNHP list of rare animal species; several are Endangered or Critically Imperiled in NYS (see attached documentation from Loyan Beausoleil’s 5-month bird survey, e- bird, and iNaturalist). All ten require thoughtful, individual mitigation plans. 2) The DEIS (6.5-13) makes unjustified assumptions about the temporary nature of negative impacts when it proposes that urban wildlife will "relocate to other suitable areas".