Redox-Based Transcriptional Regulation in Prokaryotes: Revisiting Model Mechanisms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Use of Mathematical Modeling and Other Biophysical Methods For
USE OF MATHEMATICAL MODELING AND OTHER BIOPHYSICAL METHODS FOR INSIGHTS INTO IRON-RELATED PHENOMENA OF BIOLOGICAL SYSTEMS A Dissertation by JOSHUA D. WOFFORD Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Paul A. Lindahl Committee Members, David P. Barondeau Simon W. North Vishal M. Gohil Head of Department, Simon W. North December 2018 Major Subject: Chemistry Copyright 2018 Joshua D. Wofford ABSTRACT Iron is a crucial nutrient in most living systems. It forms the active centers of many proteins that are critical for many cellular functions, either by themselves or as Fe-S clusters and hemes. However, Fe is potentially toxic to the cell in high concentrations and must be tightly regulated. There has been much work into understanding various pieces of Fe trafficking and regulation, but integrating all of this information into a coherent model has proven difficult. Past research has focused on different Fe species, including cytosolic labile Fe or mitochondrial Fe-S clusters, as being the main regulator of Fe trafficking in yeast. Our initial modeling efforts demonstrate that both cytosolic Fe and mitochondrial ISC assembly are required for proper regulation. More recent modeling efforts involved a more rigorous multi- tiered approach. Model simulations were optimized against experimental results involving respiring wild-type and Mrs3/4-deleted yeast. Simulations from both modeling studies suggest that mitochondria possess a “respiratory shield” that prevents a vicious cycle of nanoparticle formation, ISC loss, and subsequent loading of mitochondria with iron. -
Nitroxide-Mediated Polymerization
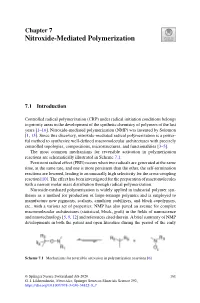
Chapter 7 Nitroxide-Mediated Polymerization 7.1 Introduction Controlled radical polymerization (CRP) under radical initiation conditions belongs to priority areas in the development of the synthetic chemistry of polymers of the last years [1–16]. Nitroxide-mediated polymerization (NMP) was invented by Solomon [1, 13]. Since this discovery, nitroxide-mediated radical polymerization is a power- ful method to synthesize well-defined macromolecular architectures with precisely controlled topologies, compositions, microstructures, and functionalities [3–5]. The most common mechanisms for reversible activation in polymerization reactions are schematically illustrated in Scheme 7.1. Persistent radical effect (PRE) occurs when two radicals are generated at the same time, at the same rate, and one is more persistent than the other, the self-termination reactions are lowered, leading to an unusually high selectivity for the cross-coupling reaction [10]. The effect has been investigated for the preparation of macromolecules with a narrow molar mass distribution through radical polymerization. Nitroxide-mediated polymerization is widely applied in industrial polymer syn- theses as a method for production of large-tonnage polymers and is employed to manufacture new pigments, sealants, emulsion stabilizers, and block copolymers, etc., with a various set of properties. NMP has also paved an avenue for complex macromolecular architectures (statistical, block, graft) in the fields of nanoscience and nanotechnology [5, 9, 12] and references cited therein. A brief summary of NMP developments in both the patent and open literature during the period of the early Scheme 7.1 Mechanisms for reversible activation in polymerization reactions [6] © Springer Nature Switzerland AG 2020 161 G. I. Likhtenshtein, Nitroxides, Springer Series in Materials Science 292, https://doi.org/10.1007/978-3-030-34822-9_7 162 7 Nitroxide-Mediated Polymerization 1980–2000 was presented in [11]. -

Electrochemical and Structural Characterization of Azotobacter Vinelandii Flavodoxin II
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Caltech Authors Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II Helen M. Segal,1 Thomas Spatzal,1 Michael G. Hill,2 Andrew K. Udit,2 and Douglas C. Rees 1* 1Division of Chemistry and Chemical Engineering, Howard Hughes Medical Institute, California Institute of Technology, Pasadena, California 91125 2Division of Chemistry, Occidental College, Los Angeles, California 90041 Received 1 June 2017; Accepted 10 July 2017 DOI: 10.1002/pro.3236 Published online 14 July 2017 proteinscience.org Abstract: Azotobacter vinelandii flavodoxin II serves as a physiological reductant of nitrogenase, the enzyme system mediating biological nitrogen fixation. Wildtype A. vinelandii flavodoxin II was electrochemically and crystallographically characterized to better understand the molecular basis for this functional role. The redox properties were monitored on surfactant-modified basal plane graphite electrodes, with two distinct redox couples measured by cyclic voltammetry correspond- ing to reduction potentials of 2483 6 1 mV and 2187 6 9 mV (vs. NHE) in 50 mM potassium phos- phate, 150 mM NaCl, pH 7.5. These redox potentials were assigned as the semiquinone/ hydroquinone couple and the quinone/semiquinone couple, respectively. This study constitutes one of the first applications of surfactant-modified basal plane graphite electrodes to characterize the redox properties of a flavodoxin, thus providing a novel electrochemical method to study this class of protein. The X-ray crystal structure of the flavodoxin purified from A. vinelandii was solved at 1.17 A˚ resolution. With this structure, the native nitrogenase electron transfer proteins have all been structurally characterized. -

Tremblay Robinlee.Pdf
Expression and Characterisation ofa Gene Enc oding RbpD, an RNA- Bind ing Protein in Anabaena sp. strain PeC 7120 by Rob in lee Tremblay A lhesis submitted to the Scltool of Graduale Studies in partial fulfilment of the requirements fOl" the degree of Master of Science Department of BiochemistrylFacultyof Science Memorial University of Newfoundland January 2000 SI.JOM'S Newfoun dland Abs t ra ct The RNA-binding protein RbpD, from the cyano bacterium Anaba ena sp, strain Pe C 7120 was expressed in £Sch~ ric h ia coli and successfully purified using me IMPACT I system (New England Biolabs). The rbp D gene was cloned into the pCYBt expre ssion vector by using polymerase chain reaction to introduce Ndel and SapI restriction sites at the 5' end 3' ends of the gene respect ively. The 3'.-end mutagenesis also chan ged the stop codon into a cysteine codon. The resulting gene encoded a fusion protein consisting of RbpD, the Saccharomyces cerev isiae VMA intein and a chitin binding domain.. Expressi on of the fusion protein was observed in £ coli strain MCI061 but Western blot analysis using an intein-directed ant ibody indicated that significant in vivo fmeln-direcred splicing of the fusion protein occurred. We were unable to eliminate this problem; no fusion protein expression was observed in 8 other E coli strains tested. Wild -type RbpD was purified following binding of the fusion protein 10 a chitin column and overnight cleavage in the presence of a reducing agent, dlthicthrehc l. A number of modifications to the manufacturer' s purification protocol were found to be necessary for success ful purification. -

Purification and Characterisation of a Protease (Tamarillin) from Tamarillo Fruit
Purification and characterisation of a protease (tamarillin) from tamarillo fruit Item Type Article Authors Li, Zhao; Scott, Ken; Hemar, Yacine; Zhang, Huoming; Otter, Don Citation Li Z, Scott K, Hemar Y, Zhang H, Otter D (2018) Purification and characterisation of a protease (tamarillin) from tamarillo fruit. Food Chemistry. Available: http://dx.doi.org/10.1016/ j.foodchem.2018.02.091. Eprint version Post-print DOI 10.1016/j.foodchem.2018.02.091 Publisher Elsevier BV Journal Food Chemistry Rights NOTICE: this is the author’s version of a work that was accepted for publication in Food Chemistry. Changes resulting from the publishing process, such as peer review, editing, corrections, structural formatting, and other quality control mechanisms may not be reflected in this document. Changes may have been made to this work since it was submitted for publication. A definitive version was subsequently published in Food Chemistry, [, , (2018-02-16)] DOI: 10.1016/j.foodchem.2018.02.091 . © 2018. This manuscript version is made available under the CC-BY-NC-ND 4.0 license http://creativecommons.org/licenses/by-nc-nd/4.0/ Download date 29/09/2021 23:19:14 Link to Item http://hdl.handle.net/10754/627180 Accepted Manuscript Purification and characterisation of a protease (tamarillin) from tamarillo fruit Zhao Li, Ken Scott, Yacine Hemar, Huoming Zhang, Don Otter PII: S0308-8146(18)30327-3 DOI: https://doi.org/10.1016/j.foodchem.2018.02.091 Reference: FOCH 22475 To appear in: Food Chemistry Received Date: 25 October 2017 Revised Date: 13 February 2018 Accepted Date: 16 February 2018 Please cite this article as: Li, Z., Scott, K., Hemar, Y., Zhang, H., Otter, D., Purification and characterisation of a protease (tamarillin) from tamarillo fruit, Food Chemistry (2018), doi: https://doi.org/10.1016/j.foodchem. -

A Mathematical Model of Iron Import and Trafficking in Wild-Type and Mrs3/4ΔΔ Yeast Cells Joshua D
Wofford and Lindahl BMC Systems Biology (2019) 13:23 https://doi.org/10.1186/s12918-019-0702-2 RESEARCH ARTICLE Open Access A mathematical model of iron import and trafficking in wild-type and Mrs3/4ΔΔ yeast cells Joshua D. Wofford1 and Paul A. Lindahl1,2* Abstract Background: Iron plays crucial roles in the metabolism of eukaryotic cells. Much iron is trafficked into mitochondria where it is used for iron-sulfur cluster assembly and heme biosynthesis. A yeast strain in which Mrs3/4, the high- affinity iron importers on the mitochondrial inner membrane, are deleted exhibits a slow-growth phenotype when grown under iron-deficient conditions. However, these cells grow at WT rates under iron-sufficient conditions. The object of this study was to develop a mathematical model that could explain this recovery on the molecular level. Results: A multi-tiered strategy was used to solve an ordinary-differential-equations-based mathematical model of iron import, trafficking, and regulation in growing Saccharomyces cerevisiae cells. At the simplest level of modeling, all iron in the cell was presumed to be a single species and the cell was considered to be a single homogeneous volume. Optimized parameters associated with the rate of iron import and the rate of dilution due to cell growth were determined. At the next level of complexity, the cell was divided into three regions, including cytosol, mitochondria, and vacuoles, each of which was presumed to contain a single form of iron. Optimized parameters associated with import into these regions were determined. At the final level of complexity, nine components were assumed within the same three cellular regions. -

ANABOLISM III: Biosynthesis Amino Acids & Nucleotides
BI/CH 422/622 ANABOLISM OUTLINE: Photosynthesis Carbohydrate Biosynthesis in Animals Biosynthesis of Fatty Acids and Lipids Biosynthesis of Amino Acids and Nucleotides Nitrogen fixation nitrogenase Nitrogen assimilation Glutamine synthetase Glutamate synthase Amino-acid Biosynthesis non-essential essential Nucleotide Biosynthesis RNA precursors purines pyrimidines DNA precursors deoxy-nucleotides Biosynthesis of secondary products of amino acids ANABOLISM III: Biosynthesis Amino Acids & Nucleotides Dr. Kornberg: Lecture 04.26.17 (0:00-5:06) 5 min 1 Biosynthesis Amino Acids & Nucleotides How are Ribonucleic Acid Precursors So far: converted to Deoxyribonucleic Acid GMPàGDPàGTP Precursors? ….....and how is dTTP made? AMPàADPàATP 2’C-OH bond is directly reduced to 2’-H UMPàUDPàUTPà bond …without activating the carbon for CDPßCTP dehydration, etc.! catalyzed by ribonucleotide reductase Specific kinases, Non-specific kinase, e.g., UMP kinase, nucleoside GMP kinase, diphosphate kinase Very unique enzyme in all of biochemistry – use of free Adenylate kinase (works on both oxy- and radicals etc. deoxy-ribose GDPàdGDP nucleosides) Mechanism: Two H atoms are donated ADPàdADP by NADPH and carried by thioredoxin or glutaredoxin to the active site. UDPàdUDP –Substrates are the NDPs and the products CDPàdCDP are dNDP. Biosynthesis Amino Acids & Nucleotides Source of Reducing Structure of Ribonucleotide Reductase a2 are regulatory Electrons for and half the Ribonucleotide catalytic site; need to be reduced. Reductase b 2 are the other half (a b ) of the active site, 2 2 and the free- radical generators • NADPH serves as the electron donor. • Funneled through glutathione or JoAnne Stubbe thioredoxin pathways (1946– ) 2 •Most forms of enzyme have two catalytic/ regulatory subunits and two radical- generating subunits. -

Discovery of Industrially Relevant Oxidoreductases
DISCOVERY OF INDUSTRIALLY RELEVANT OXIDOREDUCTASES Thesis Submitted for the Degree of Master of Science by Kezia Rajan, B.Sc. Supervised by Dr. Ciaran Fagan School of Biotechnology Dublin City University Ireland Dr. Andrew Dowd MBio Monaghan Ireland January 2020 Declaration I hereby certify that this material, which I now submit for assessment on the programme of study leading to the award of Master of Science, is entirely my own work, and that I have exercised reasonable care to ensure that the work is original, and does not to the best of my knowledge breach any law of copyright, and has not been taken from the work of others save and to the extent that such work has been cited and acknowledged within the text of my work. Signed: ID No.: 17212904 Kezia Rajan Date: 03rd January 2020 Acknowledgements I would like to thank the following: God, for sending me angels in the form of wonderful human beings over the last two years to help me with any- and everything related to my project. Dr. Ciaran Fagan and Dr. Andrew Dowd, for guiding me and always going out of their way to help me. Thank you for your patience, your advice, and thank you for constantly believing in me. I feel extremely privileged to have gotten an opportunity to work alongside both of you. Everything I’ve learnt and the passion for research that this project has sparked in me, I owe it all to you both. Although I know that words will never be enough to express my gratitude, I still want to say a huge thank you from the bottom of my heart. -

On the Natural History of Flavin-Based Electron Bifurcation
On the Natural History of Flavin-Based Electron Bifurcation Frauke Baymann, Barbara Schoepp-Cothenet, Simon Duval, Marianne Guiral, Myriam Brugna, Carole Baffert, Michael Russell, Wolfgang Nitschke To cite this version: Frauke Baymann, Barbara Schoepp-Cothenet, Simon Duval, Marianne Guiral, Myriam Brugna, et al.. On the Natural History of Flavin-Based Electron Bifurcation. Frontiers in Microbiology, Frontiers Media, 2018, 9, pp.1357 - 1357. 10.3389/fmicb.2018.01357. hal-01828959 HAL Id: hal-01828959 https://hal-amu.archives-ouvertes.fr/hal-01828959 Submitted on 5 Jul 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. fmicb-09-01357 June 29, 2018 Time: 19:12 # 1 REVIEW published: 03 July 2018 doi: 10.3389/fmicb.2018.01357 On the Natural History of Flavin-Based Electron Bifurcation Frauke Baymann1, Barbara Schoepp-Cothenet1, Simon Duval1, Marianne Guiral1, Myriam Brugna1, Carole Baffert1, Michael J. Russell2 and Wolfgang Nitschke1* 1 CNRS, BIP, UMR 7281, IMM FR3479, Aix-Marseille University, Marseille, France, 2 Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA, United States Electron bifurcation is here described as a special case of the continuum of electron transfer reactions accessible to two-electron redox compounds with redox cooperativity. -

PDF (Chapter 7)
7 Ferredoxins, Hydrogenases, and Nitrogenases: Metal-Sulfide Proteins EDWARD I. STIEFEL AND GRAHAM N. GEORGE Exxon Research and Engineering Company Transition-metal/sulfide sites, especially those containing iron, are present in all forms of life and are found at the active centers of a wide variety of redox and catalytic proteins. These proteins include simple soluble electron-transfer agents (the ferredoxins), membrane-bound components of electron-transfer chains, and some of the most complex metalloenzymes, such as nitrogenase, hydrogenase, and xanthine oxidase. In this chapter we first review the chemistry of the Fe-S sites that occur in relatively simple rubredoxins and ferredoxins, and make note of the ubiquity of these sites in other metalloenzymes. We use these relatively simple systems to show the usefulness of spectroscopy and model-system studies for deducing bioinorganic structure and reactivity. We then direct our attention to the hydro genase and nitrogenase enzyme systems, both of which use transition-metal sulfur clusters to activate and evolve molecular hydrogen. I. IRON-SULFUR PROTEINS AND MODELS Iron sulfide proteins involved in electron transfer are called ferredoxins and rub redoxins. * The ferredoxins were discovered first, and were originally classified as bacterial (containing Fe4S4 clusters) and plant (containing FezSz clusters) fer redoxins. This classification is now recognized as being not generally useful, since both FezSz and Fe4S4 ferredoxins are found in plants,14,15 animals, Z,6,16 and bacteria.4 Ferredoxins are distinguished from rubredoxins by their posses sion of acid-labile sulfide; i.e., an inorganic Sz- ion that forms HzS gas upon denaturation at low pH. -

Electron Transfer and Substrate Reduction in Nitrogenase
Utah State University DigitalCommons@USU All Graduate Theses and Dissertations Graduate Studies 5-2014 Electron Transfer and Substrate Reduction in Nitrogenase Karamatullah Danyal Utah State University Follow this and additional works at: https://digitalcommons.usu.edu/etd Part of the Biochemistry Commons Recommended Citation Danyal, Karamatullah, "Electron Transfer and Substrate Reduction in Nitrogenase" (2014). All Graduate Theses and Dissertations. 2181. https://digitalcommons.usu.edu/etd/2181 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@USU. It has been accepted for inclusion in All Graduate Theses and Dissertations by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. ELECTRON TRANSFER AND SUBSTRATE REDUCTON IN NITROGENASE by Karamatullah Danyal A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biochemistry Approved: ________________________ _______________________ Lance C. Seefeldt Scott A. Ensign Major Professor Committee Member ________________________ _______________________ Alvan C. Hengge Sean J. Johnson Committee Member Committee Member ________________________ _______________________ Korry Hintze Mark R. McLellan Committee Member Vice President for Research and Dean of the School of Graduate Studies UTAH STATE UNIVERSITY Logan, Utah 2014 ii Copyright © Karamatullah Danyal 2014 All Rights Reserved iii ABSTRACT Electron Transfer and Substrate Reduction in Nitrogenase by Karamatullah Danyal, Doctor of Philosophy Utah State University, 2014 Major Professor: Dr. Lance C. Seefeldt Department: Chemistry and Biochemistry Population growth over the past ~50 years accompanied by the changes in dietary habits due to economic growth have markedly increased the demand for fixed nitrogen. Aided by biological nitrogen fixation, the Haber-Bosch process has been able to fulfill these demands. -

Molybdenum Hazards to Fish, Wildlife, and Invertebrates: a Synoptic Review
Biological Report 85(1.19) Contaminant Hazard Reviews August 1989 Report No. 19 MOLYBDENUM HAZARDS TO FISH, WILDLIFE, AND INVERTEBRATES: A SYNOPTIC REVIEW by Ronald Eisler U.S. Fish and Wildlife Service Patuxent Wildlife Research Center Laurel, MD 20708 SUMMARY The element molybdenum (Mo) is found in all living organisms and is considered to be an essential or beneficial micronutrient. However, the molybdenum poisoning of ruminants has been reported in at least 15 States and 8 foreign countries. Molybdenum is used primarily in the manufacture of steel alloys. Its residues tend to be elevated in plants and soils near Mo mining and reclamation sites, fossil-fuel power plants, and Mo disposal areas. Concentrations of Mo are usually lower in fish and wildlife than in terrestrial macrophytes. Aquatic organisms are comparatively resistant to Mo salts: adverse effects on growth and survival usually appeared only at water concentrations >50 mg Mo/l. But in one study, 50% of newly fertilized eggs of rainbow trout (Oncorhynchus mykiss) died in 28 days at only 0.79 mg Mo/l. High bioconcentration of Mo by certain species of aquatic algae and invertebrates--up to 20 grams of Mo/kg dry weight--has been recorded without apparent harm to the accumulator; however, hazard potential to upper trophic organisms (such as waterfowl) that may feed on bioconcentrators is not clear. Data on Mo effects are missing for avian wildlife and are inadequate for mammalian wildlife. In domestic birds, adverse effects on growth have been reported at dietary Mo concentrations of 200 mg Mo/kg, on reproduction at 500 mg/kg, and on survival at 6,000 mg/kg.