Computing the Diameter of a Point Set Grégoire Malandain, Jean-Daniel Boissonnat
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

P. 1 Math 490 Notes 7 Zero Dimensional Spaces for (SΩ,Τo)
p. 1 Math 490 Notes 7 Zero Dimensional Spaces For (SΩ, τo), discussed in our last set of notes, we can describe a basis B for τo as follows: B = {[λ, λ] ¯ λ is a non-limit ordinal } ∪ {[µ + 1, λ] ¯ λ is a limit ordinal and µ < λ}. ¯ ¯ The sets in B are τo-open, since they form a basis for the order topology, but they are also closed by the previous Prop N7.1 from our last set of notes. Sets which are simultaneously open and closed relative to the same topology are called clopen sets. A topology with a basis of clopen sets is defined to be zero-dimensional. As we have just discussed, (SΩ, τ0) is zero-dimensional, as are the discrete and indiscrete topologies on any set. It can also be shown that the Sorgenfrey line (R, τs) is zero-dimensional. Recall that a basis for τs is B = {[a, b) ¯ a, b ∈R and a < b}. It is easy to show that each set [a, b) is clopen relative to τs: ¯ each [a, b) itself is τs-open by definition of τs, and the complement of [a, b)is(−∞,a)∪[b, ∞), which can be written as [ ¡[a − n, a) ∪ [b, b + n)¢, and is therefore open. n∈N Closures and Interiors of Sets As you may know from studying analysis, subsets are frequently neither open nor closed. However, for any subset A in a topological space, there is a certain closed set A and a certain open set Ao associated with A in a natural way: Clτ A = A = \{B ¯ B is closed and A ⊆ B} (Closure of A) ¯ o Iτ A = A = [{U ¯ U is open and U ⊆ A}. -

Molecular Symmetry
Molecular Symmetry Symmetry helps us understand molecular structure, some chemical properties, and characteristics of physical properties (spectroscopy) – used with group theory to predict vibrational spectra for the identification of molecular shape, and as a tool for understanding electronic structure and bonding. Symmetrical : implies the species possesses a number of indistinguishable configurations. 1 Group Theory : mathematical treatment of symmetry. symmetry operation – an operation performed on an object which leaves it in a configuration that is indistinguishable from, and superimposable on, the original configuration. symmetry elements – the points, lines, or planes to which a symmetry operation is carried out. Element Operation Symbol Identity Identity E Symmetry plane Reflection in the plane σ Inversion center Inversion of a point x,y,z to -x,-y,-z i Proper axis Rotation by (360/n)° Cn 1. Rotation by (360/n)° Improper axis S 2. Reflection in plane perpendicular to rotation axis n Proper axes of rotation (C n) Rotation with respect to a line (axis of rotation). •Cn is a rotation of (360/n)°. •C2 = 180° rotation, C 3 = 120° rotation, C 4 = 90° rotation, C 5 = 72° rotation, C 6 = 60° rotation… •Each rotation brings you to an indistinguishable state from the original. However, rotation by 90° about the same axis does not give back the identical molecule. XeF 4 is square planar. Therefore H 2O does NOT possess It has four different C 2 axes. a C 4 symmetry axis. A C 4 axis out of the page is called the principle axis because it has the largest n . By convention, the principle axis is in the z-direction 2 3 Reflection through a planes of symmetry (mirror plane) If reflection of all parts of a molecule through a plane produced an indistinguishable configuration, the symmetry element is called a mirror plane or plane of symmetry . -

The Motion of Point Particles in Curved Spacetime
Living Rev. Relativity, 14, (2011), 7 LIVINGREVIEWS http://www.livingreviews.org/lrr-2011-7 (Update of lrr-2004-6) in relativity The Motion of Point Particles in Curved Spacetime Eric Poisson Department of Physics, University of Guelph, Guelph, Ontario, Canada N1G 2W1 email: [email protected] http://www.physics.uoguelph.ca/ Adam Pound Department of Physics, University of Guelph, Guelph, Ontario, Canada N1G 2W1 email: [email protected] Ian Vega Department of Physics, University of Guelph, Guelph, Ontario, Canada N1G 2W1 email: [email protected] Accepted on 23 August 2011 Published on 29 September 2011 Abstract This review is concerned with the motion of a point scalar charge, a point electric charge, and a point mass in a specified background spacetime. In each of the three cases the particle produces a field that behaves as outgoing radiation in the wave zone, and therefore removes energy from the particle. In the near zone the field acts on the particle and gives rise toa self-force that prevents the particle from moving on a geodesic of the background spacetime. The self-force contains both conservative and dissipative terms, and the latter are responsible for the radiation reaction. The work done by the self-force matches the energy radiated away by the particle. The field’s action on the particle is difficult to calculate because of its singular nature:the field diverges at the position of the particle. But it is possible to isolate the field’s singular part and show that it exerts no force on the particle { its only effect is to contribute to the particle's inertia. -
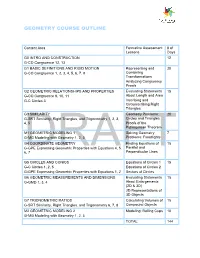
Geometry Course Outline
GEOMETRY COURSE OUTLINE Content Area Formative Assessment # of Lessons Days G0 INTRO AND CONSTRUCTION 12 G-CO Congruence 12, 13 G1 BASIC DEFINITIONS AND RIGID MOTION Representing and 20 G-CO Congruence 1, 2, 3, 4, 5, 6, 7, 8 Combining Transformations Analyzing Congruency Proofs G2 GEOMETRIC RELATIONSHIPS AND PROPERTIES Evaluating Statements 15 G-CO Congruence 9, 10, 11 About Length and Area G-C Circles 3 Inscribing and Circumscribing Right Triangles G3 SIMILARITY Geometry Problems: 20 G-SRT Similarity, Right Triangles, and Trigonometry 1, 2, 3, Circles and Triangles 4, 5 Proofs of the Pythagorean Theorem M1 GEOMETRIC MODELING 1 Solving Geometry 7 G-MG Modeling with Geometry 1, 2, 3 Problems: Floodlights G4 COORDINATE GEOMETRY Finding Equations of 15 G-GPE Expressing Geometric Properties with Equations 4, 5, Parallel and 6, 7 Perpendicular Lines G5 CIRCLES AND CONICS Equations of Circles 1 15 G-C Circles 1, 2, 5 Equations of Circles 2 G-GPE Expressing Geometric Properties with Equations 1, 2 Sectors of Circles G6 GEOMETRIC MEASUREMENTS AND DIMENSIONS Evaluating Statements 15 G-GMD 1, 3, 4 About Enlargements (2D & 3D) 2D Representations of 3D Objects G7 TRIONOMETRIC RATIOS Calculating Volumes of 15 G-SRT Similarity, Right Triangles, and Trigonometry 6, 7, 8 Compound Objects M2 GEOMETRIC MODELING 2 Modeling: Rolling Cups 10 G-MG Modeling with Geometry 1, 2, 3 TOTAL: 144 HIGH SCHOOL OVERVIEW Algebra 1 Geometry Algebra 2 A0 Introduction G0 Introduction and A0 Introduction Construction A1 Modeling With Functions G1 Basic Definitions and Rigid -

Name: Introduction to Geometry: Points, Lines and Planes Euclid
Name: Introduction to Geometry: Points, Lines and Planes Euclid: “What’s the point of Geometry?- Euclid” http://www.youtube.com/watch?v=_KUGLOiZyK8&safe=active When do historians believe that Euclid completed his work? What key topic did Euclid build on to create all of Geometry? What is an axiom? Why do you think that Euclid’s ideas have lasted for thousands of years? *Point – is a ________ in space. A point has ___ dimension. A point is name by a single capital letter. (ex) *Line – consists of an __________ number of points which extend in ___________ directions. A line is ___-dimensional. A line may be named by using a single lower case letter OR by any two points on the line. (ex) *Plane – consists of an __________ number of points which form a flat surface that extends in all directions. A plane is _____-dimensional. A plane may be named using a single capital letter OR by using three non- collinear points. (ex) *Line segment – consists of ____ points on a line, called the endpoints and all of the points between them. (ex) *Ray – consists of ___ point on a line (called an endpoint or the initial point) and all of the points on one side of the point. (ex) *Opposite rays – share the same initial point and extend in opposite directions on the same line. *Collinear points – (ex) *Coplanar points – (ex) *Two or more geometric figures intersect if they have one or more points in common. The intersection of the figures is the set of elements that the figures have in common. -

1 Line Symmetry
Name: ______________________________________________________________ Date: _________________________ Period: _____ Chapter 4: Transformations Topic 10: Symmetry There are three types of symmetry that we will consider. #1 Line Symmetry: A line of symmetry not only cuts a figure in___________________, it creates a mirror image. In order to determine if a figure has line symmetry, a figure needs to be able to be divided into two “mirror image halves.” The line of symmetry is ____________________________from all corresponding pairs of points. Another way to think about line symmetry: If the image is reflected of the line of symmetry, it will return the same image. Examples: In the figures below, sketch all the lines of symmetry (if any): Think critically: Some have one, some have many, some have none! Remember, if the image is reflected over the line you draw, it should be identical to how it started! More Examples: Letters and numbers can also have lines of symmetry! Sketch as many lines of symmetry as you can (if any): Name: ______________________________________________________________ Date: _________________________ Period: _____ #2 Rotational Symmetry: RECALL! The total measure of degrees around a point is: _____________ A rotational symmetry of a figure is a rotation of the plane that maps the figure back to itself such that the rotation is greater than , but less than ___________________. In regular polygons (polygons in which all sides are congruent), the number of rotational symmetries equals the number of sides of the figure. If a polygon is not regular, it may have fewer rotational symmetries. We can consider increments of like coordinate plane rotations. Examples of Regular Polygons: A rotation of will always map a figure back onto itself. -

Points, Lines, and Planes a Point Is a Position in Space. a Point Has No Length Or Width Or Thickness
Points, Lines, and Planes A Point is a position in space. A point has no length or width or thickness. A point in geometry is represented by a dot. To name a point, we usually use a (capital) letter. A A (straight) line has length but no width or thickness. A line is understood to extend indefinitely to both sides. It does not have a beginning or end. A B C D A line consists of infinitely many points. The four points A, B, C, D are all on the same line. Postulate: Two points determine a line. We name a line by using any two points on the line, so the above line can be named as any of the following: ! ! ! ! ! AB BC AC AD CD Any three or more points that are on the same line are called colinear points. In the above, points A; B; C; D are all colinear. A Ray is part of a line that has a beginning point, and extends indefinitely to one direction. A B C D A ray is named by using its beginning point with another point it contains. −! −! −−! −−! In the above, ray AB is the same ray as AC or AD. But ray BD is not the same −−! ray as AD. A (line) segment is a finite part of a line between two points, called its end points. A segment has a finite length. A B C D B C In the above, segment AD is not the same as segment BC Segment Addition Postulate: In a line segment, if points A; B; C are colinear and point B is between point A and point C, then AB + BC = AC You may look at the plus sign, +, as adding the length of the segments as numbers. -

Lecture 2: Analyzing Algorithms: the 2-D Maxima Problem
Lecture Notes CMSC 251 Lecture 2: Analyzing Algorithms: The 2-d Maxima Problem (Thursday, Jan 29, 1998) Read: Chapter 1 in CLR. Analyzing Algorithms: In order to design good algorithms, we must first agree the criteria for measuring algorithms. The emphasis in this course will be on the design of efficient algorithm, and hence we will measure algorithms in terms of the amount of computational resources that the algorithm requires. These resources include mostly running time and memory. Depending on the application, there may be other elements that are taken into account, such as the number disk accesses in a database program or the communication bandwidth in a networking application. In practice there are many issues that need to be considered in the design algorithms. These include issues such as the ease of debugging and maintaining the final software through its life-cycle. Also, one of the luxuries we will have in this course is to be able to assume that we are given a clean, fully- specified mathematical description of the computational problem. In practice, this is often not the case, and the algorithm must be designed subject to only partial knowledge of the final specifications. Thus, in practice it is often necessary to design algorithms that are simple, and easily modified if problem parameters and specifications are slightly modified. Fortunately, most of the algorithms that we will discuss in this class are quite simple, and are easy to modify subject to small problem variations. Model of Computation: Another goal that we will have in this course is that our analyses be as independent as possible of the variations in machine, operating system, compiler, or programming language. -

Impact of Geometer's Sketchpad on Students Achievement in Graph
The Malaysian Online Journal of Educational Technology Volume 1, Issue2 Impact Of Geometer’s Sketchpad On [1] Faculty of Education University of Malaya Students Achievement In Graph Functions 50603 Kuala Lumpur, Malaysia [email protected] Leong Kwan Eu [1] ABSTRACT The purpose of this study is to investigate the effect of using the Geometer’s Sketchpad software in the teaching and learning of graph functions among Form Six (Grade 12) students in a Malaysian secondary school. This study utilized a quasi-experimental design using intact group of students from two classes in an urban secondary school. Two instruments were used to gather information on the students’ mathematics achievement and attitudes towards learning graph functions.A survey questionnaire was used to gauge students’ perception on the usage of Geometer’s Sketchpad in the learning of graph functions.In the study, the experimental groups used Geometer’s Sketchpad-based worksheets and each student has an access to one computer equipped with the Sketchpad software. The control group used only the textbooks. Both groups took the same pre- test.This study indicated that the results ofthe experimental group had a significant mean difference compared to the control group. The use of Geometer’s Sketchpad in mathematics classroom has positive effect on the students’ mathematics achievement and attitude towards the learning of graph of functions also improved. Graph Functions, students achievement, dynamical Keywords: geometric software, educational technology, students attitude INTRODUCTION Geometers’ Sketchpad encourage and facilitate those new broader conceptions of school algebra, and they raise deep questions about the appropriate role of traditional symbol manipulation skills (CBMS, 2000). -

LECTURE 3: LINES and PLANES in THREE-DIMENSIONAL SPACE 1. Lines in R We Can Now Use Our New Concepts of Dot Products and Cross P
LECTURE 3: LINES AND PLANES IN THREE-DIMENSIONAL SPACE MA1111: LINEAR ALGEBRA I, MICHAELMAS 2016 1. Lines in R3 We can now use our new concepts of dot products and cross products to describe some examples of writing down geometric objects efficiently. We begin with the question of determining equations for lines in R3, namely, real three-dimensional space (like the world we live in). Aside. Since it is inconvenient to constantly say that v is an n-dimensional vector, etc., we will start using the notation Rn for n-dimensional vectors. For example, R1 is the real-number line, and R2 is the xy-plane. Our methods will of course work in the plane, R2, as well. Instead of beginning with a formal statement, we motivate the general idea by beginning with an example. Every line is determined by a point on the line and a direction. For example, suppose we want to write an equation for the line passing through the point A = (1; 3; 5) and which is parallel to the vector (2; 6; 7). Note that if I took any non-zero multiple of v, the following argument would work equally well. Now suppose that B = (x; y; z) is an arbitrary point. Then −! −! B is on L () AB points in the direction of v () AB = tv; t 2 R 8 x = 1 + 2t <> () (x − 1; y − 3; z − 5) = (2t; 6t; 7t) () y = 3 + 6t for t 2 R: :>z = 5 + 7t These are the parametric equations for L. The general formula isn't much harder to derive. -

A Geometric Introduction to Spacetime and Special Relativity
A GEOMETRIC INTRODUCTION TO SPACETIME AND SPECIAL RELATIVITY. WILLIAM K. ZIEMER Abstract. A narrative of special relativity meant for graduate students in mathematics or physics. The presentation builds upon the geometry of space- time; not the explicit axioms of Einstein, which are consequences of the geom- etry. 1. Introduction Einstein was deeply intuitive, and used many thought experiments to derive the behavior of relativity. Most introductions to special relativity follow this path; taking the reader down the same road Einstein travelled, using his axioms and modifying Newtonian physics. The problem with this approach is that the reader falls into the same pits that Einstein fell into. There is a large difference in the way Einstein approached relativity in 1905 versus 1912. I will use the 1912 version, a geometric spacetime approach, where the differences between Newtonian physics and relativity are encoded into the geometry of how space and time are modeled. I believe that understanding the differences in the underlying geometries gives a more direct path to understanding relativity. Comparing Newtonian physics with relativity (the physics of Einstein), there is essentially one difference in their basic axioms, but they have far-reaching im- plications in how the theories describe the rules by which the world works. The difference is the treatment of time. The question, \Which is farther away from you: a ball 1 foot away from your hand right now, or a ball that is in your hand 1 minute from now?" has no answer in Newtonian physics, since there is no mechanism for contrasting spatial distance with temporal distance. -

12 Notation and List of Symbols
12 Notation and List of Symbols Alphabetic ℵ0, ℵ1, ℵ2,... cardinality of infinite sets ,r,s,... lines are usually denoted by lowercase italics Latin letters A,B,C,... points are usually denoted by uppercase Latin letters π,ρ,σ,... planes and surfaces are usually denoted by lowercase Greek letters N positive integers; natural numbers R real numbers C complex numbers 5 real part of a complex number 6 imaginary part of a complex number R2 Euclidean 2-dimensional plane (or space) P2 projective 2-dimensional plane (or space) RN Euclidean N-dimensional space PN projective N-dimensional space Symbols General ∈ belongs to such that ∃ there exists ∀ for all ⊂ subset of ∪ set union ∩ set intersection A. Inselberg, Parallel Coordinates, DOI 10.1007/978-0-387-68628-8, © Springer Science+Business Media, LLC 2009 532 Notation and List of Symbols ∅ empty st (null set) 2A power set of A, the set of all subsets of A ip inflection point of a curve n principal normal vector to a space curve nˆ unit normal vector to a surface t vector tangent to a space curve b binormal vector τ torsion (measures “non-planarity” of a space curve) × cross product → mapping symbol A → B correspondence (also used for duality) ⇒ implies ⇔ if and only if <, > less than, greater than |a| absolute value of a |b| length of vector b norm, length L1 L1 norm L L norm, Euclidean distance 2 2 summation Jp set of integers modulo p with the operations + and × ≈ approximately equal = not equal parallel ⊥ perpendicular, orthogonal (,,) homogeneous coordinates — ordered triple representing