Multidirectional Chromosome Painting Substantiates the Occurrence of Extensive Genomic Reshuffling Within Accipitriformes Wenhui Nie1*, Patricia C
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Griffon Vultures &Lpar;<I>Gyps Fulvus</I>&Rpar; Ingesting Bones At
SEPTEMBER1997 LETTERS 287 AravaipaCreek, Arizona. Almost immediatelyat leastsix Elf Owls beganvocalizing from dispersedlocations around our campsiteand at leastfour of them began making low passesat the Great Horned Owl. Before we could relocate the horned owl to a protectedenclosure, it wasstruck once in the head by an Elf Owl. Some nocturnal behaviorsmay not be well known or understood,not becausethey are rare, but becausethey are difficult to observe.This may changewith the increasedavailability of night vision equipment (P. Henson and J A Cooper 1994, Auk 111:1013-1018). Currently, observationsof nocturnal behaviorsare likely to be sporadicand anecdotal,and therefore unreported. Such information, however,may help in understandinga speciesbiology. For example, other researchershave observedgroup mobbing by Elf Owls (F.R. Gehlbach,pers. comm.; B.A. Millsap, pers. comm.), but there are no publishedreports of the behavior.Our observations,and thoseof other researchers, suggestthat Elf Owlswill join together in mobbing and that they can be physicallyaggressive when defending their nestsagainst predators. We thank A. Duerr, T.S. Estabrookand R.L. Spauldingfor assistingwith the observations.We alsothank T. Brush, ER. Gehlbach, R. Glinski, P. Hardy, B.A. Millsap, G. Proudfoot and H.A. Snyder for sharing their observational information concerning mobbing by small owls.This manuscriptbenefitted from the constructivereviews of ER. Gehlbach,C. Marti, B.A. Millsapand an anonymousreviewer.--Clint W. Boal, Brent D. Biblesand R. William Mannan, Schoolof RenewableNatural Resources,University of Arizona, Tucson,AZ 85721 U.S•. j. RaptorRes. 31 (3):287-288 ¸ 1997 The Raptor ResearchFoundation, Inc. GRIFFONVULTURES (GYPS FULVUS) INGESTING BONES AT THE OSSUARIESOF BEARDEDVULTURES ( GYPAETUSBAR•ATUS) Some African vulturesovercome the calcium deficiencyin their diets by ingestingbone fragments,and are depen- dent on the presenceof largepredators to supplythem (Mundy and Ledger 1976, S. -

Disaggregation of Bird Families Listed on Cms Appendix Ii
Convention on the Conservation of Migratory Species of Wild Animals 2nd Meeting of the Sessional Committee of the CMS Scientific Council (ScC-SC2) Bonn, Germany, 10 – 14 July 2017 UNEP/CMS/ScC-SC2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II (Prepared by the Appointed Councillors for Birds) Summary: The first meeting of the Sessional Committee of the Scientific Council identified the adoption of a new standard reference for avian taxonomy as an opportunity to disaggregate the higher-level taxa listed on Appendix II and to identify those that are considered to be migratory species and that have an unfavourable conservation status. The current paper presents an initial analysis of the higher-level disaggregation using the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World Volumes 1 and 2 taxonomy, and identifies the challenges in completing the analysis to identify all of the migratory species and the corresponding Range States. The document has been prepared by the COP Appointed Scientific Councilors for Birds. This is a supplementary paper to COP document UNEP/CMS/COP12/Doc.25.3 on Taxonomy and Nomenclature UNEP/CMS/ScC-Sc2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II 1. Through Resolution 11.19, the Conference of Parties adopted as the standard reference for bird taxonomy and nomenclature for Non-Passerine species the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World, Volume 1: Non-Passerines, by Josep del Hoyo and Nigel J. Collar (2014); 2. -

Chromosome Painting in Three Species of Buteoninae: a Cytogenetic Signature Reinforces the Monophyly of South American Species
Chromosome Painting in Three Species of Buteoninae: A Cytogenetic Signature Reinforces the Monophyly of South American Species Edivaldo Herculano C. de Oliveira1,2,3*, Marcella Mergulha˜o Tagliarini4, Michelly S. dos Santos5, Patricia C. M. O’Brien3, Malcolm A. Ferguson-Smith3 1 Laborato´rio de Cultura de Tecidos e Citogene´tica, SAMAM, Instituto Evandro Chagas, Ananindeua, PA, Brazil, 2 Faculdade de Cieˆncias Exatas e Naturais, ICEN, Universidade Federal do Para´, Bele´m, PA, Brazil, 3 Cambridge Resource Centre for Comparative Genomics, Cambridge, United Kingdom, 4 Programa de Po´s Graduac¸a˜oem Neurocieˆncias e Biologia Celular, ICB, Universidade Federal do Para´, Bele´m, PA, Brazil, 5 PIBIC – Universidade Federal do Para´, Bele´m, PA, Brazil Abstract Buteoninae (Falconiformes, Accipitridae) consist of the widely distributed genus Buteo, and several closely related species in a group called ‘‘sub-buteonine hawks’’, such as Buteogallus, Parabuteo, Asturina, Leucopternis and Busarellus, with unsolved phylogenetic relationships. Diploid number ranges between 2n = 66 and 2n = 68. Only one species, L. albicollis had its karyotype analyzed by molecular cytogenetics. The aim of this study was to present chromosomal analysis of three species of Buteoninae: Rupornis magnirostris, Asturina nitida and Buteogallus meridionallis using fluorescence in situ hybridization (FISH) experiments with telomeric and rDNA probes, as well as whole chromosome probes derived from Gallus gallus and Leucopternis albicollis. The three species analyzed herein showed similar karyotypes, with 2n = 68. Telomeric probes showed some interstitial telomeric sequences, which could be resulted by fusion processes occurred in the chromosomal evolution of the group, including the one found in the tassociation GGA1p/GGA6. -

Vulture Msap)
MULTI-SPECIES ACTION PLAN TO CONSERVE AFRICAN-EURASIAN VULTURES (VULTURE MSAP) CMS Raptors MOU Technical Publication No. 5 CMS Technical Series No. xx MULTI-SPECIES ACTION PLAN TO CONSERVE AFRICAN-EURASIAN VULTURES (VULTURE MSAP) CMS Raptors MOU Technical Publication No. 5 CMS Technical Series No. xx Overall project management Nick P. Williams, CMS Raptors MOU Head of the Coordinating Unit [email protected] Jenny Renell, CMS Raptors MOU Associate Programme Officer [email protected] Compiled by André Botha, Endangered Wildlife Trust Overarching Coordinator: Multi-species Action Plan to conserve African-Eurasian Vultures [email protected] Jovan Andevski, Vulture Conservation Foundation European Regional Coordinator: Multi-species Action Plan to conserve African-Eurasian Vultures [email protected] Chris Bowden, Royal Society for the Protection of Birds Asian Regional Coordinator: Multi-species Action Plan to conserve African-Eurasian Vultures [email protected] Masumi Gudka, BirdLife International African Regional Coordinator: Multi-species Action Plan to conserve African-Eurasian Vultures [email protected] Roger Safford, BirdLife International Senior Programme Manager: Preventing Extinctions [email protected] Nick P. Williams, CMS Raptors MOU Head of the Coordinating Unit [email protected] Technical support Roger Safford, BirdLife International José Tavares, Vulture Conservation Foundation Regional Workshop Facilitators Africa - Chris Bowden, Royal Society for the Protection of Birds Europe – Boris Barov, BirdLife International Asia and Middle East - José Tavares, Vulture Conservation Foundation Overarching Workshop Chair Fernando Spina, Chair of the CMS Scientific Council Design and layout Tris Allinson, BirdLife International 2 Multi-species Action Plan to Conserve African-Eurasian Vultures (Vulture MsAP) Contributors Lists of participants at the five workshops and of other contributors can be found in Annex 1. -

The Reintroduction of Eurasian Griffon Monachus Vultures to France
Chancellor, R. D. & B.-U. Meyburg eds. 2004 Raptors Worldwide WWGBP/MME A Success Story: The Reintroduction of Eurasian Griffon Gyps fulvus and Black Aegypius monachus Vultures to France Michel Terrasse, François Sarrazin, Jean-Pierre Choisy, Céline Clémente, Sylvain Henriquet, Philippe Lécuyer, Jean Louis Pinna, and Christian Tessier ABSTRACT By the end of the 1960s, the future of vultures in France appeared bleak. Apart from the western half of the Pyrenees, with residual populations of Eurasian Griffon Gyps fulvus and Bearded Vultures Gypaetus barbatus, and Corsica with a few Bearded Vultures, most of France had lost its large vultures including the Black Vulture Aegypius monachus. Within the scope of a national conservation campaign, a process to restore raptor communities began. Concerning vultures, this was completed through reintroduction programmes. After the success of the Griffon Vulture reintroduction, started in 1968 in the Grands Causses of the Massif Central, other programmes followed in the 90s: Black Vultures in the Massif Central and then Griffon Vultures in the Southern Alps. In 2003 a viable population of Griffon Vultures in the Massif Central contained around 110 pairs. The same situation occurred in the Alps with about 50 breeding pairs of Griffon Vultures. Ten years after the beginning of the Black Vulture releases, the free ranging population included 11 breeding pairs. Accurate monitoring during the reintroduction period allowed us to estimate demographic parameters such as survival and breeding rates, evolution of breeding and foraging territories, main threats, movements between reintroduced populations and those from neighbouring countries, acceptance by people and the beneficial part played by vultures in what is called sustainable development. -

Spizaetus Boletin De La Red De Rapaces Neotropicales
SPIZAETUS BOLETIN DE LA RED DE RAPACES NEOTROPICALES NÚMERO 27 JUNIO 2019 BUTEO PLATYPTERUS EN COSTA RICA MILVAGO CHIMACHIMA EN COSTA RICA ASIO STYGIUS EN COLOMBIA BUTEO BRACHYURUS EN MÉXICO FALCO FEMORALIS EN MÉXICO SPIZAETUS BOLETIN DE LA RRN Número 27 © Junio 2019 Edición en Español, ISSN 2157-8966 Foto de la Portada: Juvenil de Gavilán Aludo (Buteo platypterus) en Costa Rica © Víctor Acosta-Chaves Traductores/Editores: Laura Andréa Lindenmeyer de Sousa & Marta Curti Diseño Gráfico: Marta Curti Spizaetus: Bolettin de la Red de Rapaces Neotropicales © Junio 2019 www.neotropicalraptors.org Este boletín puede ser reproducido, descargado y distribuido por fines no comerciales. Para volvera publicar cualquier artículo que figuran en este documento, por favor póngase en contacto con los autores correspondientes Contenido Singular comportamiento del caracara cabecigualdo (MILVAGO CHIMACHIMA: Falconidae) en el sur de Costa Rica José Manuel Mora & Estefanía González ............................................................................2 Nuevas localidades en Colombia del Búho Negruzco (ASIO STYGIUS) Elvis Felipe Quintero Quintero, Jeyson Sanabria-Mejía, Angélica Magaly Sogamoso Hernández & Sergio Chaparro-Herrera .................................................................................................................9 Un caso de agresión de BUTEO BRACHYURUS contra PSEUDASTUR ALBICOLLIS (Accipi- triformes) en el sur de Huimanguillo, Tabasco, México Saúl Sánchez Soto .......................................................................................................13 -

Trinidad & Tobago 2018 Species List
Trinidad Tobago Leader: Ernesto Carman Eagle-Eye Tours Nov 29 - Dec 9, 2018 Bird Species Seen/ Common Name Scientific Name Heard TINAMOUS 1 Little Tinamou Crypturellus soui H GUANS, CHACHALACAS, AND CURASSOWS 2 Trinidad Piping-Guan Aburria pipile S 3 Rufous-vented Chachalaca Ortalis ruficauda S FLAMINGOS 4 American Flamingo Phoenicopterus ruber S TROPICBIRDS 5 Red-billed Tropicbird Phaethon aethereus S FRIGATEBIRDS 6 Magnificent Frigatebird Fregata magnificens S BOOBIES AND GANNETS 7 Brown Booby Sula leucogaster S 8 Red-footed Booby Sula sula S CORMORANTS AND SHAGS 9 Neotropic Cormorant Phalacrocorax brasilianus S ANHINGAS 10 Anhinga Anhinga anhinga S PELICANS 11 Brown Pelican Pelecanus occidentalis S HERONS, EGRETS, AND BITTERNS 12 Pinnated Bittern Botaurus pinnatus S 13 Great Egret Ardea alba S 14 Little Egret Egretta garzetta S 15 Snowy Egret Egretta thula S 16 Little Blue Heron Egretta caerulea S 17 Tricolored Heron Egretta tricolor S 18 Cattle Egret Bubulcus ibis S 19 Green Heron Butorides virescens S 20 Striated Heron Butorides striata S 21 Black-crowned Night-Heron Nycticorax nycticorax S 22 Yellow-crowned Night-Heron Nyctanassa violacea S 23 Boat-billed Heron Cochlearius cochlearius S IBISES AND SPOONBILLS 24 Scarlet Ibis Eudocimus ruber S NEW WORLD VULTURES Page 1 of 8 www.eagle-eye.com Trinidad Tobago Leader: Ernesto Carman Eagle-Eye Tours Nov 29 - Dec 9, 2018 Bird Species Seen/ Common Name Scientific Name Heard 25 Black Vulture Coragyps atratus S 26 Turkey Vulture Cathartes aura S OSPREY 27 Osprey Pandion haliaetus S HAWKS, -
Avian Taxonomy
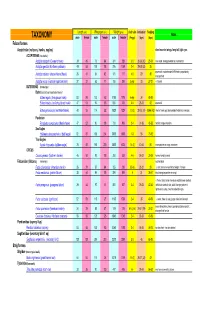
Length (cm) Wing span (cm) Weight (gms) cluch size incubation fledging Notes TAXONOMY male female male female male female (# eggs) (days) (days) Falconiformes Accipitridae (vultures, hawks, eagles) short rounded wings; long tail; light eyes ACCIPITRINAE (true hawks) Accipiter cooperii (Cooper's hawk) 39 45 73 84 341 528 3-5 30-36 (30) 25-34 crow sized; strongly banded tail; rounded tail Accipiter gentilis (Northern goshawk) 49 58 101 108 816 1059 2-4 28-38 (33) 35 square tail; most abundant NAM hawk; proportionaly 26 31 54 62 101 177 4-5 29 30 Accipiter striatus (sharp-shinned hawk) strongest foot Accipiter nisus (Eurasian sparrowhawk ) 37 37 62 77 150 290 5-Apr 33 27-31 Musket BUTEONINAE (broadwings) Buteo (buzzards or broad tailed hawks) Buteo regalis (ferruginous hawk) 53 59 132 143 1180 1578 6-Apr 34 45-50 Buteo lineatus (red-shouldered hawk) 47 53 96 105 550 700 3-4 28-33 42 square tail Buteo jamaicensis (red-tailed hawk) 48 55 114 122 1028 1224 1-3 (3) 28-35 (34) 42-46 (42) (Harlan' hawk spp); dark patagial featehres: immature; Parabuteo Parabuteo cuncincutus (Harris hawk) 47 52 90 108 710 890 2-4 31-36 45-50 reddish orange shoulders Sea Eagles Haliaeetus leucocephalus (bald eagle) 82 87 185 244 3000 6300 1-3 35 70-92 True Eagles Aquila chrysaetos (golden eagle) 78 82 185 220 3000 6125 1-4 (2) 40-45 50 white patches on wings: immature; CIRCUS Circus cyaneus (Northern harrier) 46 50 93 108 350 530 4-6 26-32 30-35 hovers; hunts by sound Falconidae (falcons) (longwings) notched beak Falco columbarius (American merlin) 26 29 57 64 -

Recent Literature and Book Reviews
Vulture News 69 November 2015 RECENT LITERATURE AND BOOK REVIEWS P.J. Mundy BAMFORD, A. J., MONADJEM, A., DIEKMANN, M. & HARDY, I. C.W. (2009). Development of non-explosive-based methods for mass capture of vultures. South African Journal of Wildlife Research 39: 202-208. Investigated methods of launching nets, powered by black powder, detonators, elastic, and compressed air, and a walk-in trap. The last proved most effective at capturing birds, needed no licence, but took a long time to construct and of course was barely portable. (email for Monadjem: [email protected]) BIRD, J. P. & BLACKBURN, T. M. (2011). Observations of large raptors in northeast Sudan. Scopus 31: 19-27. From a 430km transect (Atbara to Port Sudan) and other ad hoc observations, in January 2010, three species of vulture were seen. Thirty Egyptian Vultures on the transect, counts of 21 and 24 Lappet-faced Vultures at a carcass and one Hooded Vulture, were noted. (email: [email protected]) BREWSTER, C. A & TYLER, S. J. (2012). Summary of category B records. Babbler 57: 45-47. Many sightings are listed for the Cape, Lappet-faced, White-headed and Hooded Vultures, but no ages. In the Tswapong Hills, no nests of the Cape Vulture were seen at Lerala, but 76 were seen at Moremi Gorge. On p. 57 many sightings are listed of the White-backed Vulture, from singles up to groups of about 70. c ( /o BLB, Box 26691, Gaborone) 51 Vulture News 69 November 2015 BRUCE-MILLER, I. (2008). Martial Eagle Polemaetus bellicosus apparently killing White-headed Vulture Trigonoceps occipitalis. -

Trade of Threatened Vultures and Other Raptors for Fetish and Bushmeat in West and Central Africa
Trade of threatened vultures and other raptors for fetish and bushmeat in West and Central Africa R. BUIJ,G.NIKOLAUS,R.WHYTOCK,D.J.INGRAM and D . O GADA Abstract Diurnal raptors have declined significantly in Introduction western Africa since the s. To evaluate the impact of traditional medicine and bushmeat trade on raptors, we ex- ildlife is exploited throughout West and Central amined carcasses offered at markets at sites (– stands WAfrica (Martin, ). The trade in bushmeat (wild- per site) in countries in western Africa during –. sourced meat) has been recognized as having a negative im- Black kite Milvus migrans andhoodedvultureNecrosyrtes pact on wildlife populations in forests and savannahs monachus together accounted for %of, carcasses com- (Njiforti, ; Fa et al., ; Lindsey et al., ), and it . prising species. Twenty-seven percent of carcasses were of is estimated billion kg of wild animal meat is traded an- species categorized as Near Threatened, Vulnerable or nually in Central Africa (Wilkie & Carpenter, ). In add- Endangered on the IUCN Red List. Common species were ition to being exploited for bushmeat, many animals are traded more frequently than rarer species, as were species hunted and traded specifically for use in traditional medi- with frequent scavenging behaviour (vs non-scavenging), gen- cine, also known as wudu, juju or fetish (Adeola, ). eralist or savannah habitat use (vs forest), and an Afrotropical The use of wildlife items for the treatment of a range of (vs Palearctic) breeding range. Large Afrotropical vultures physical and mental diseases, or to bring good fortune, were recorded in the highest absolute and relative numbers has been reported in almost every country in West and in Nigeria, whereas in Central Africa, palm-nut vultures Central Africa. -

Lista Oficial PN
Nombre científico Inglés Español Crypturellus souiC Little Tinamou Tinamú Chico Crypturellus cinnamomeusR Thicket Tinamou Tinamú Canelo Dendrocygna autumnalis Black-bellied Whistling-Duck Pijije Común Cairina moschata Muscovy Duck Pato Real Ortalis vetula Plain Chachalaca Chachalaca Olivácea Penelope purpurascens Crested Guan Pava Crestada Crax rubra Great Curassow Pavón Grande Colinus cristatus Crested Bobwhite Codorniz Crestada Tachybaptus dominicus Least Grebe Zambullidor Enano Ardenna creatopus Pink-footed Shearwater Pardela Blanca Común Ardenna pacificus Wedge-tailed Shearwater Pardela Colicuña Ardenna grisea Sooty Shearwater Pardela Sombría Puffinus nativitatis Christmas Shearwater Pardela de Navidad, Pardela de Christmas Puffinus subalaris Galapagos Shearwater Pardela de las Galápagos Puffinus opisthomelas Black-vented Shearwater Pardela Culinegra Oceanodroma leucorhoa Leach's Storm-Petrel Paiño de Leach Oceanodroma tethys Wedge-rumped Storm-Petrel Paiño Danzarin Oceanodroma melania Black Storm-Petrel Paiño Negro Oceanodroma microsoma Least Storm-Petrel Paiño Menudo Phaethon aethereus Red-billed Tropicbird Rabijunco Piquirrojo Mycteria americana Wood Stork Cigüeñón Fregata magnificens Magnificent Frigatebird Rabihorcado Magno Sula dactylatra Masked Booby Piquero Blanco Sula granti Nazca Booby Piquero de Nazca AOCR Sula nebouxiiC Blue-footed Booby Piquero Patiazul Sula variegta Peruvian Booby Piquero Peruano Sula leucogasterR Brown Booby Piquero Moreno Sula sula Red-footed Booby Piquero Patirrojo Phalacrocorax brasilianus Neotropic -

SMCSP & SMCSN Wildlife List.Xlsx
Appendix C: Wildlife list for Six Mile Cypress Slough and Six Mile Cypress North Preserves Designated Status Scientific Name Common Name FWC FWS FNAI MAMMALS Family: Didelphidae (opossums) Didelphis virginiana Virginia opossum Family: Dasypodidae (armadillos) Dasypus novemcinctus nine-banded armadillo * Family: Sciuridae (squirrels and their allies) Sciurus carolinensis eastern gray squirrel Sciurus niger avicennia Big Cypress fox squirrel T G5T2/S2 Family: Muridae (mice and rats) Peromyscus gossypinus cotton mouse Oryzomys palustris marsh rice rat Sigmodon hispidus hispid cotton rat Family: Leporidae (rabbits and hares) Sylvilagus palustris marsh rabbit Sylvilagus floridanus eastern cottontail Family: Talpidae (moles) Scalopus aquaticus eastern mole Family: Felidae (cats) Puma concolor coryi Florida panther E E G5T1/S1 Lynx rufus bobcat Felis silvestris domestic cat * Family: Canidae (wolves and foxes) Urocyon cinereoargenteus common gray fox Family: Ursidae (bears) Ursus americanus floridanus Florida black bear T G5T2/S2 Family: Procyonidae (raccoons) Procyon lotor raccoon Family: Mephitidae (skunks) Spilogale putorius eastern spotted skunk Mephitis mephitis striped skunk Family: Mustelidae (weasels, otters and relatives) Lutra canadensis northern river otter Family: Suidae (old world swine) Sus scrofa feral hog * Family: Cervidae (deer) Odocoileus virginianus white-tailed deer BIRDS Family: Anatidae (swans, geese and ducks) Subfamily: Anatinae (dabbling ducks) Dendrocygna autumnalis black-bellied whistling duck Cairina moschata muscovy