Faisal10.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download PDF # Sylviidae: Chrysomma, Fulvetta, Paradoxornis
NINOGGSXCNNZ \\ Doc > Sylviidae: Chrysomma, Fulvetta, Paradoxornis, Parisoma, Sylvia, Parrotbill, Old World warbler, Jerdon's Babbler,... Sylviidae: Chrysomma, Fulvetta, Paradoxornis, Parisoma, Sylvia, Parrotbill, Old World warbler, Jerdon's Babbler, Wrentit, Blackcap Filesize: 6.59 MB Reviews Very beneficial to all type of folks. I could comprehended every thing using this created e pdf. I found out this book from my i and dad suggested this book to find out. (Ms. Madaline Nienow) DISCLAIMER | DMCA GPPUH64MHWS0 < eBook \\ Sylviidae: Chrysomma, Fulvetta, Paradoxornis, Parisoma, Sylvia, Parrotbill, Old World warbler, Jerdon's Babbler,... SYLVIIDAE: CHRYSOMMA, FULVETTA, PARADOXORNIS, PARISOMA, SYLVIA, PARROTBILL, OLD WORLD WARBLER, JERDON'S BABBLER, WRENTIT, BLACKCAP To download Sylviidae: Chrysomma, Fulvetta, Paradoxornis, Parisoma, Sylvia, Parrotbill, Old World warbler, Jerdon's Babbler, Wrentit, Blackcap PDF, please access the hyperlink listed below and save the document or have access to additional information that are have conjunction with SYLVIIDAE: CHRYSOMMA, FULVETTA, PARADOXORNIS, PARISOMA, SYLVIA, PARROTBILL, OLD WORLD WARBLER, JERDON'S BABBLER, WRENTIT, BLACKCAP ebook. Books LLC, Wiki Series, 2016. Paperback. Book Condition: New. PRINT ON DEMAND Book; New; Publication Year 2016; Not Signed; Fast Shipping from the UK. No. book. Read Sylviidae: Chrysomma, Fulvetta, Paradoxornis, Parisoma, Sylvia, Parrotbill, Old World warbler, Jerdon's Babbler, Wrentit, Blackcap Online Download PDF Sylviidae: Chrysomma, Fulvetta, Paradoxornis, Parisoma, -

Thick-Billed Warbler (Iduna Aedon) at Gambell, Alaska: First Record for North America Gary H
NOTES THICK-BILLED WARBLER (IDUNA AEDON) AT GAMBELL, ALASKA: FIRST RECORD FOR NORTH AMERICA GARY H. ROSENBERG, 8101 North Wheatfield Dr., Tucson, Arizona 85741; [email protected] PAUL E. LEHMAN, 11192 Portobelo Dr., San Diego, California 92124; [email protected] AARON J. LANG, 40208 Alpenglow Circle, Homer, Alaska 99603; [email protected] VICTOR AND RUBEN STOLL, 899 Miller Rd., Centerville, Tennessee 37033; [email protected] In the evening on 8 September 2017, in the “far boneyard” at Gambell, St. Law- rence Island, Alaska (63.78° N, 171.74° W), Victor and Ruben Stoll flushed a pas- serine they could not immediately identify. The “boneyards” are large pits excavated by the resident Yupik Natives seeking buried ivory and artifacts, a result of several thousand years of sea-mammal hunting from this island’s Northwest Cape. Working these pits turns the soil, which has resulted in the growth of relatively lush vegetation consisting of two species of Artemisia, known locally as “wormwood.” The combina- tion of lush vegetation (reaching 0.5–1 m in height) and deep depressions that offer protection from the wind is attractive to migrant and vagrant landbirds in the otherwise flat, gravelly landscape. Soon thereafter, we, along with Greg Scyphers, Monte Taylor, and other birders then at Gambell, converged at the far boneyard in search of the bird. It was soon relocated and seen on the ground briefly by Lang, who suggested it was a Thick-billed Warbler (Iduna aedon), a bird he was familiar with from southeastern Asia and a species not previously recorded in Alaska or North America. -

Based on Nuclear and Mitochondrial Sequence Data
MOLECULAR PHYLOGENETICS AND EVOLUTION Molecular Phylogenetics and Evolution 29 (2003) 126–138 www.elsevier.com/locate/ympev Phylogeny of Passerida (Aves: Passeriformes) based on nuclear and mitochondrial sequence data Per G.P. Ericsona,* and Ulf S. Johanssona,b a Department of Vertebrate Zoology and Molecular Systematics Laboratory, Swedish Museum of Natural History, Frescativagen 44, P.O. Box 50007, SE-10405 Stockholm, Sweden b Department of Zoology, University of Stockholm, SE-106 91 Stockholm, Sweden Received 18 September 2002; revised 23 January 2003 Abstract Passerida is a monophyletic group of oscine passerines that includes almost 3500 species (about 36%) of all bird species in the world. The current understanding of higher-level relationships within Passerida is based on DNA–DNA hybridizations [C.G. Sibley, J.E. Ahlquist, Phylogeny and Classification of Birds, 1990, Yale University Press, New Haven, CT]. Our results are based on analyses of 3130 aligned nucleotide sequence data obtained from 48 ingroup and 13 outgroup genera. Three nuclear genes were sequenced: c-myc (498–510 bp), RAG-1 (930 bp), and myoglobin (693–722 bp), as well one mitochondrial gene; cytochrome b (879 bp). The data were analysed by parsimony, maximum-likelihood, and Bayesian inference. The African rockfowl and rock- jumper are found to constitute the deepest branch within Passerida, but relationships among the other taxa are poorly resolved— only four major clades receive statistical support. One clade corresponds to Passeroidea of [C.G. Sibley, B.L. Monroe, Distribution and Taxonomy of Birds of the World, 1990, Yale University Press, New Haven, CT] and includes, e.g., flowerpeckers, sunbirds, accentors, weavers, estrilds, wagtails, finches, and sparrows. -

Composition of Bird Species in Huu Lien Nature Reserve, Lang Son Province
VNU Journal of Science, Earth Sciences 27 (2011) 13-24 Composition of bird species in Huu Lien Nature Reserve, Lang Son province Nguyen Lan Hung Son*, Nguyen Thi Hoa, Le Trung Dung Hanoi National University of Education, 136 Xuan Thuy, Hanoi, Vietnam Received 3 December 2010; received in revised form 17 December 2010 Abstract . The diversity of bird species is of special importance as it can create responsive and adaptive behaviours among the whole animal population in wild environment. For this reason, the frequent making of inventory lists of bird species helps assess and evaluate the current status of forest resources in natural conservation areas which are inherently under human pressures in our country. During the two years (2009 - 2010) of the study conducted in Huu Lien Nature Reserve in Lang Son province, records have been made of 168 bird species belonging to 117 genus, 54 families, 17 orders. Of these, 9 bird species are rare and of high value of genetic preservation. Discussions have been held on the data for classification and arrangement of bird lists. This regional avifauna is characterized as typical of the lime stone mountain ecology in the Northeast of Vietnam along the border with China. The illegal activities of timbering Buretiodendron hsienmu take place at high frequency are making it a threat to the conservation of the diversity of bird species in this area. Keywords: avifauna, lime stone mountain, rare species, timbering. 1. Introduction ∗∗∗ hectares and an buffer zone of another 10.000 hectares. On May 31 st 2006, the Chairman of Huu Lien Nature Reserve was recognized as Lang Son People’s Committee issued the in the Decision numbered 194/CT dated August decision numbered 705/Q Đ-UBND on th 9 1986 by the Council of Ministers. -

14Th of November 2019
Date: 14th of november 2019 Tour: Half day excursion Oostvaardersplassen Guide: Pim Around 8.30 I met Quentin from Vancouver (Canada) at the small trainstation of Almere Centrum. From here we drove to the nature reserve Oostvaardersplassen. Our first stop was at a viewing point. From a distance we saw a large group of Red deer, on farmland we saw a few smaller Roe deer. We saw our first White tailed eagle of the day, his nickname is “flying door”, because his wingspan is about 2.30 meters! At the moment there are about 10-12 of this large birds in the area and we saw this morning half of this population. In The Netherlands there are about 10 breeding pairs, it al started in the Oostvaardersplassen about 13 years ago. We saw many Common buzzard, Common kestrel and a male Eurasian sparrowhaw was showing well closeby. We saw a large group of Barnacle geese and Greylag geese and a mixed group of hundreds of Northern lapwings and European golden plovers. Also a adult Whooper swan was present. Eurasian sparrowhawk – Oostvaardersplassen – Lelystad. “The whooper swan is a large white swan, bigger than a Bewick's swan. It has a long thin neck, which it usually holds erect, and black legs. Its black bill has a large triangular patch of yellow on it”. Source: Rspb.org. The Whooper swans we saw in the reserve today are mainly from Russia and Finland, a pair stays together for the rest of their live On our second stop, also a viewing point, we saw a juvenile White tailed eagle and two adults in a tree. -

AIRONE CENERINO Ardea Cinerea the Grey Heron (Ardea Cinerea
AIRONE CENERINO Ardea cinerea The Grey Heron (Ardea cinerea ) is a wading bird of the heron family Ardeidae . The Grey Heron is a large bird, standing 1 m tall, and it has a 1.5 m wingspan. It is the largest European heron. Its plumage is largely grey above, and off-white below. It has a powerful yellow bill, which is brighter in breeding adults. It has a slow flight, with its long neck retracted (S-shaped). This is characteristic of herons and bitterns, and distinguishes them from storks, cranes and spoonbills, which extend their necks This species breeds in colonies in trees close to lakes or other wetlands, although it will also nest in reed beds. It builds a bulky stick nest. It feeds in shallow water, spearing fish or frogs with its long, sharp bill. Herons will also take small mammals and birds. It will often wait motionless for prey, or slowly stalk its victim. The call is a loud croaking "fraaank". This species is very similar to the American Great Blue Heron. The Australian White-faced Heron is often incorrectly called Grey Heron ALLODOLA Alauda arvensis The Skylark (Alauda arvensis ) is a small passerine bird. It breeds across most of Europe and Asia and in the mountains of north Africa. It is mainly resident in the west of its range, but eastern populations of are more migratory, moving further south in winter. Even in the milder west of its range, many birds move to lowlands and the coast in winter. Asian birds appear as vagrants in Alaska; this bird has also been introduced in Hawaii and western North America. -

This Dissertation Has Been 61-3328 Microfilmed Exactly As Received
THE TAXONOMIC SIGNIFICANCE OF THE TONGUE MUSCULATURE OF PASSERINE BIRDS Item Type text; Dissertation-Reproduction (electronic) Authors George, William Gordon, 1925- Publisher The University of Arizona. Rights Copyright © is held by the author. Digital access to this material is made possible by the University Libraries, University of Arizona. Further transmission, reproduction or presentation (such as public display or performance) of protected items is prohibited except with permission of the author. Download date 05/10/2021 15:17:28 Link to Item http://hdl.handle.net/10150/284412 This dissertation has been 61-3328 microfilmed exactly as received GEORGE, William Gordon, 1925- THE TAXONOMIC SIGNIFICANCE OF THE TONGUE MUSCULATURE OF PASSERINE BIRDS. University of Arizona, Ph.D., 1961 Zoology University Microfilms, Inc., Ann Arbor, Michigan THE TAXONOMIC SIGNIFICANCE OF THE TONGUE MUSCULATURE OF PASSERINE BIRDS by William G. George A Dissertation Submitted to the Faculty of the DEPARTMENT OF ZOOLOGY In Partial Fulfillment of the Requirements For the Degree of DOCTOR OF PHILOSOPHY In the Graduate College THE UNIVERSITY OF ARIZONA 1961 THE UNIVERSITY OF ARIZONA GRADUATE COLLEGE I hereby recommend that this dissertation prepared under my direction by Wllllaa G. George entitled The Taxonoalc Significance of the Tongue Musculature of Passerine Birds be accepted as fulfilling the dissertation requirement of the degree of Doctor of Philosophy Dissertation Director / Date After inspection of the dissertation, the following members of the Final Examination Committee concur in its approval and recommend its acceptance:* *This approval and acceptance Is contingent on the candidate's adequate performance and defense of this dissertation at the final oral examination. -
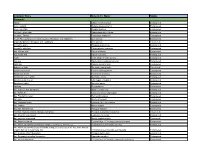
List of Endangered Species
Common Name Scienticfic Name Status Mammals Addax Addax nasomaculatus Endangered Anoa, lowland Bubalus depressicornis Endangered Anoa, mountain Bubalus quarlesi Endangered Antelope, giant sable Hippotragus niger variani Endangered Antelope, Tibetan Panthalops hodgsonii Endangered Argali [All populations except Kyrgyzstan, Mongolia, and Tajikistan] Ovis ammon Endangered Argali [Kyrgyzstan, Mongolia, and Tajikistan] Ovis ammon Threatened Armadillo, giant Priodontes maximus Endangered Armadillo, pink fairy Chlamyphorus truncatus Endangered Ass, African wild Equus africanus Endangered Ass, Asian wild Equus hemionus Endangered Avahi Avahi laniger(=entire genus) Endangered Aye-aye Daubentonia madagascariensis Endangered Babirusa Babyrousa babyrussa Endangered Baboon, gelada Theropithecus gelada Threatened Bandicoot, barred Perameles bougainville Endangered Bandicoot, desert Perameles eremiana Endangered Bandicoot, lesser rabbit Macrotis leucura Endangered Bandicoot, pig-footed Chaeropus ecaudatus Endangered Bandicoot, rabbit Macrotis lagotis Endangered Banteng Bos javanicus Endangered Bat, Bulmer's fruit (flying fox) Aproteles bulmerae Endangered Bat, bumblebee Craseonycteris thonglongyai Endangered Bat, Florida bonneted Eumops floridanus Endangered Bat, gray Myotis grisescens Endangered Bat, Hawaiian hoary Lasiurus cinereus semotus Endangered Bat, Indiana Myotis sodalis Endangered Bat, little Mariana fruit Pteropus tokudae Endangered Fruit Bat, Mariana (=fanihi, Mariana flying fox) Pteropus mariannus mariannus Threatened Bat, Mexican long-nosed -

Ph.D. Og Speciale
UNIVERSITY OF COPENH AGEN FACULTY OR DEPARTMENT Space use strategies of two Palaearctic migrants on the wintering grounds A study of winter territoriality in the Common Chiffchaff and Subalpine Warbler Dina Abdelhafez Ali Mostafa Supervisor: Kasper Thorup Co-Supervisor: Torben Dabelsteen Submitted on: 2 October, 2017 Name of department: Natural History Museum of Denmark Behavioural Ecology - Section of Ecology and Evolution - Department of Biology. University of Copenhagen Author(s): Dina Abdelhafez Ali Mostafa Title and subtitle: Space use strategies of two Palaearctic migrants on the wintering grounds : A study of winter territoriality in the Common Chiffchaff and Subalpine Warbler Topic description: An investigation of the chosen space use strategies and territoriality in two migrant songbirds wintering in the Sahel. Supervisor: Kasper Thorup – Torben Dablesteen Submitted on: 2 October 2017 Grade: Number of study units: 2 3 2 Table of contents PREFACE ............................................................................................................................ 4 SUMMARY .......................................................................................................................... 7 BACKGROUND .................................................................................................................. 9 Why study birds on their winter ground ...................................................................................................................... 10 Sociality as an extension of space use strategies -

Western Birds Journal
WESTERN BIRDS Vol. 49, No. 3, 2018 Western Specialty: Black Swift Photo by © Commander Michael G. Levine, NOAA ship Oscar Dyson: Nazca Booby (Sula granti) ~27 km south of the southern tip of the Kenai Peninsula, Alaska, 30 August 2017 The Nazca Booby nests principally on the Galapagos Islands and on Malpelo Island off Colombia. It was not confirmed to disperse north as far as the U.S. until 2013, but since Photo by © Sue Hirshman of Montrose, Colorado: then over a dozen are known to have reached California. In 2017 one strayed as far north Black Swift (Cypseloides niger) even as the western margin of the Gulf of Alaska, as reported in this issue of Western Box Canyon, Ouray, Colorado, 31 July 2013 Birds by Daniel D. Gibson, Lucas H. DeCicco, Robert E. Gill Jr., Steven C. Heinl, Aaron J. Lang, Theodore J. Tobish Jr., and Jack J. Withrow in the fourth report of the One of North America’s most difficult birds to study, the Black Swift has long kept many Alaska Checklist Committee. secrets. Among these are the extent of the species’ sexual dimorphism and how its plumage may change over time. In this issue of Western Birds, Carolyn Gunn, Kevin J. Aagaard, This incursion of the Nazca Booby thousands of miles from its normal range may Kim M. Potter, and Jason P. Beason report the answers to these questions on the basis of represent bad news for the species. In a study published in 2017 recapturing adults at their nest sites over a period of 14 years and sexing them by genetic (PLoS One 12[8]:e0182545), Emily M. -

11. Birds of the Paradise Gardens
Mute Swan Cygnus olor The mute swan is a species of swan, and thus a member of the waterfowl family Anatidae. It is native to much of Europe and Asia, and the far north of Africa. It is an introduced species in North America, Australasia and southern Africa Tundra Swan Cygnus columbianus The tundra swan is a small Holarctic swan. The two taxa within it are usually regarded as conspecific, but are also sometimes split into two species: Bewick's swan of the Palaearctic and the whistling swan proper of the Nearctic Bean Goose Anser fabalis The bean goose is a goose that breeds in northern Europe and Asia. It has two distinct varieties, one inhabiting taiga habitats and one inhabiting tundra Red-breasted Goose Branta ruficollis The red-breasted goose is a brightly marked species of goose in the genus Branta from Eurasia. It is sometimes separated in Rufibrenta but appears close enough to the brant goose to make this unnecessary, despite its distinct appearance Common Shelduck Tadorna tadorna The common shelduck is a waterfowl species shelduck genus Tadorna. It is widespread and common in Eurasia, mainly breeding in temperate and wintering in subtropical regions; in winter, it can also be found in the Maghreb Eurasian Teal Anas crecca The Eurasian teal or common teal is a common and widespread duck which breeds in temperate Eurasia and migrates south in winter Mallard Anas platyrhynchos The mallard or wild duck is a dabbling duck which breeds throughout the temperate and subtropical Americas, Europe, Asia, and North Africa, and has been introduced to New Zealand, Australia, Peru, .. -

Clay Pit Ponds Bird Checklist
Observer Field Data Sp Su F W Sp Su F W Sp Su F W NOTES: MOCKINGBIRDS, NAME _________________________________ THRASHERS, & ALLIES __Northern Waterthrush U U __Common Grackle* CCCU DATE ______________ TIME ______________ __Gray Catbird* C C C R __Connecticut Warbler R __Brown-headed Cowbird* UUUR BIRDOF S __Northern Mockingbird* C C C C __Mourning Warbler R __Orchard Oriole R WEATHER _____________________________ __Brown Thrasher* C C C R __Common Yellowthroat* C C C __Baltimore Oriole* U U ________________________________________ C L A Y P I T P O N D S __Hooded Warbler R S T A T E P A R K P R E S E R V E STARLINGS AND ALLIES __Wilson’s Warbler U FRINGILLINE & CARDUELINE FINCHES ________________________________________ __European Starling* C C C C __Canada Warbler U U __Purple Finch R R NOTES ________________________________ __Yellow-breasted Chat # R R __House Finch* CCCC _________________________________________ WAXWINGS __Common Redpoll R R __Cedar Waxwing U U R TANAGERS __Pine Siskin R R ________________________________________ __Summer Tanager R __American Goldfinch* UUUU Special observations (breeding behavior, rare or out of WOOD WARBLERS __Scarlet Tanager U U __Evening Grosbeak R R season sightings, etc.) should be reported to: __Blue-winged Warbler U R Clay Pit Ponds State Park Preserve __Golden-winged Warbler # R NEW WORLD SPARROWS OLD WORLD SPARROWS 83 Nielsen Avenue __Tennessee Warbler R R __Eastern Towhee* C C U R __House Sparrow* CCCC Staten Island, NY 10309 __Orange-crowned Warbler R __American Tree Sparrow U (718) 967-1976