Expression Patterns of Gmap2/EREB-Like Transcription Factors Involved in Soybean Responses to Water Deficit
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Investigation of Selected Wood Properties and the Suitability for Industrial Utilization of Acacia Seyal Var
Fakultät Umweltwissenschaften - Faculty of Environmental Sciences Investigation of selected wood properties and the suitability for industrial utilization of Acacia seyal var. seyal Del and Balanites aegyptiaca (L.) Delile grown in different climatic zones of Sudan Dissertation to achieve the academic title Doctor rerum silvaticarum (Dr. rer. silv.) Submitted by MSc. Hanadi Mohamed Shawgi Gamal born 23.09.1979 in Khartoum/Sudan Referees: Prof. Dr. Dr. habil. Claus-Thomas Bues, Dresden University of Technology Prof Dr. Andreas Roloff, Dresden University of Technology Prof. Dr. Dr. h.c. František Hapla, University of Göttingen Dresden, 07.02.2014 1 Acknowledgement Acknowledgement First of all, I wish to praise and thank the god for giving me the strength and facilitating things throughout my study. I would like to express my deep gratitude and thanks to Prof. Dr. Dr. habil. Claus-Thomas Bues , Chair of Forest Utilization, Institute of Forest Utilization and Forest Technology, Dresden University of Technology, for his continuous supervision, guidance, patience, suggestion, expertise and research facilities during my research periods in Dresden. He was my Father and Supervisor, I got many advices from his side in the scientific aspect as well as the social aspects. He taught me how to be a good researcher and gives me the keys of the wood science. I am grateful to Dr. rer. silv. Björn Günther for his help and useful comments throughout the study. He guides me in almost all steps in my study. Dr.-Ing. Michael Rosenthal was also a good guide provided many advices, thanks for him. I would also like to express my thanks to Frau Antje Jesiorski, Frau Liane Stirl, Dipl.- Forsting. -
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
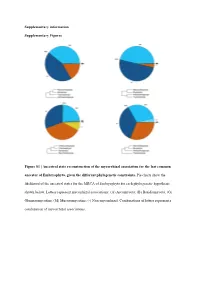
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Plastome Evolution and a Novel Loss of the Inverted Repeat In
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.04.429812; this version posted February 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The chicken or the egg? Plastome evolution and a novel loss of the inverted repeat in papilionoid legumes. Chaehee Lee1, In-Su Choi2, Domingos Cardoso3, Haroldo C. de Lima4, Luciano P. de Queiroz5, Martin F. Wojciechowski2, Robert K. Jansen1,6, and Tracey A Ruhlman1* 1Department of Integrative Biology, University of Texas at Austin 2School of Life Sciences, Arizona State University, Tempe, Arizona 85287-4501 USA 3Instituto de Biologia, Universidade Federal de Bahia (UFBA), Rua Barão de Jeremoabo, s.n., Ondina, 40170-115, Salvador, Bahia, Brazil 4Instituto de Pesquisas Jardim Botânico do Rio de Janeiro, Rua Pacheco Leão, 915 22460-030, Rio de Janeiro, Brazil 5Universidade Estadual de Feira de Santana, Av. Transnordestina, s/n, Novo Horizonte 44036-900, Feira de Santana, Bahia, Brazil 6Center of Excellence for Bionanoscience Research, King Abdulaziz University (KAU), Jeddah, Saudi Arabia *For correspondence (email [email protected]) bioRxiv preprint doi: https://doi.org/10.1101/2021.02.04.429812; this version posted February 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -

Expression Patterns of Gmap2/EREB-Like Transcription Factors Involved in Soybean Responses to Water Deficit
Expression Patterns of GmAP2/EREB-Like Transcription Factors Involved in Soybean Responses to Water Deficit Juliana Marcolino-Gomes1,2, Fabiana Aparecida Rodrigues2, Maria Cristina Neves Oliveira2, Jose Renato Bouc¸as Farias2, Norman Neumaier2, Ricardo Vilela Abdelnoor2, Francismar Correˆa Marcelino- Guimara˜es2, Alexandre Lima Nepomuceno2,3* 1 Department of Biology, State University of Londrina, Londrina, Brazil, 2 Brazilian Enterprise for Agricultural Research–Embrapa Soybean, Londrina, Brazil, 3 Embrapa LABEX US Plant Biotechnology at ARS/USDA Plant Gene Expression Center, Albany, New York, United States of America Abstract Soybean farming has faced several losses in productivity due to drought events in the last few decades. However, plants have molecular mechanisms to prevent and protect against water deficit injuries, and transcription factors play an important role in triggering different defense mechanisms. Understanding the expression patterns of transcription factors in response to water deficit and to environmental diurnal changes is very important for unveiling water deficit stress tolerance mechanisms. Here, we analyzed the expression patterns of ten APETALA2/Ethylene Responsive Element Binding-like (AP2/ EREB-like) transcription factors in two soybean genotypes (BR16: drought-sensitive; and Embrapa 48: drought-tolerant). According to phylogenetic and domain analyses, these genes can be included in the DREB and ERF subfamilies. We also analyzed a GmDRIP-like gene that encodes a DREB negative regulator. We detected the up-regulation of 9 GmAP2/EREB-like genes and identified transcriptional differences that were dependent on the levels of the stress applied and the tissue type analyzed (the expression of the GmDREB1F-like gene, for example, was four times higher in roots than in leaves). -

Seed Germination of Caragana Species from Different Regions Is
www.nature.com/scientificreports OPEN Seed germination of Caragana species from diferent regions is strongly driven by environmental Received: 16 May 2017 Accepted: 22 August 2017 cues and not phylogenetic signals Published: xx xx xxxx Xiang-Wen Fang1, Juan-Juan Zhang1, Dang-Hui Xu1, Jiayin Pang2, Tian-Peng Gao3, Chun-Hui Zhang4, Feng-Min Li1 & Neil C. Turner 1,2 Seed germination behavior is an important factor in the distribution of species. Many studies have shown that germination is controlled by phylogenetic constraints, however, it is not clear whether phylogenetic constraints or environmental cues explain seed germination of a genus from a common ancestor. In this study, seed germination under diferent temperature- and water-regimes [induced by diferent osmotic potentials of polyethylene glycol (PEG)] was investigated in the phylogenetically- related Caragana species that thrive in arid, semiarid, semihumid and humid environments. The results showed that the fnal percentage germination (FPG) decreased from 95% in species from arid habitats to 0% in species from humid habitats, but with no signifcant phylogenetic signal. Rather, the response of seed germination to temperature and PEG varied greatly with species from arid to humid habitats and was tightly linked to the ecological niche of the species, their seed coat structure and abscisic acid concentration. The fndings are not consistent with the hypothesis that within a family or a genus, seed germination strategies can be a stable evolutionary trait, thus constraining interspecifc variation, but the results clearly show that seed germination of Caragana species distributed across a range of habitats has adapted to the environment of that habitat. -

Adaptation of Growth and Respiration of Three Varieties of Caragana to Environmental Temperature
Asian Journal of Plant Sciences 7 (I): 67-72, 2008 ISSN 1682-3974 O 2008 Asian Network for Scientific Information Adaptation of Growth and Respiration of Three Varieties of Caragana to Environmental Temperature 'Weili Yu, 3Lee D. Hansen, 'Wenying Fan, 'Wenyi Zhao and 4E.Durant McArthur 'College of Soil and Water Conservation, Beijing Forest University, Beijing 100083, China 'Inner Mongolia Academy of Forest and Science, Huhhot, Inner Mongolia 010010, China 3Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah 84602, USA 4USDA Forest Service, Rocky Mountain Research Station, Shrub Sciences Laboratory, Provo, Utah 84606, USA Abstract: Growth and respiratoIy characteristics of Caragana korshinskii from Wushen and two dfferent seed sources of C. davazamcii from Hehnger and Yimhuole, all grown at the same conditions, were determined by measuring metabolic heat and CO, production rates by isothermal calorimetq at 5°C intervals from 10 to 40°C. Substrate carbon conversion efficiencies and growth (anabolic) rates were calculated from the measured data. Differences in substrate carbon conversion efficiency, respiratoIy rates and temperature responses of respiratoIy rates among the three accessiom all contribute to produce dffering temperature responses of growth rate. Planting seeds from these seed sources outside their native ranges will probably not be successful because of the differences in temperature adaptation. Key words: Wushen Caragana korshinskii, Yijmhuole Caragana davazamcii, Helinger Caragana davazamcii, substrate carbon conversion efficiency, CO, production rate, metabolic heat rate, INTRODUCTION vegetation. C. korshinskii is a species of the southern Alashan and western Ordos where it occurs in fixed and Temperature is one of the most significant semi-fixed dunes of the steppe desert and the typical determinants of plant distribution. -

2017 RBG Kew Science Publications Abreu*, JA, Hawkins, JA, Cotrim, H
2017 RBG Kew Science Publications Abreu*, J.A., Hawkins, J.A., Cotrim, H., Fay*, M.F., Hidalgo*, O. & Pellicer*, J. (2017). Ophrys fusca and Ophrys dyris (Orchidaceae) – constancy of tetraploidy amongst populations in Central Portugal. New Journal of Botany 7: 94–100. DOI: 10.1080/20423489.2017.1408185. Available online Ainsworth*, A.M. (2017). Nomenclatural novelties. Index Fungorum 332 Alderton, E., Sayer, C.D., Davies*, R., Lambert, A.J. & Axmacher, J.C. (2017). Buried alive: Aquatic plants survive in ‘ghost ponds’ under agricultural fields. Biological Conservation 212 (A): 105–110. DOI: 10.1016/j.biocon.2017.06.004. Available online Allkin*, R., Patmore*, K., Black*, N., Booker, A., Canteiro*, C., Dauncey, E., Edwards*, S., Forest*, F., Giovannini*, P., Howes*, M.-J.R., Hudson*, A., Irving*, J., Leon*, C., Milliken*, W., Nic Lughadha*, E., Schippmann, U. & Simmonds*, M.S.J. (2017). Useful plants – medicines. In Willis*, K. J. (ed) State of the World’s Plants 2017. Report. Richmond: Royal Botanic Gardens, Kew. 22–29 pp. Álvarez-Lafuente, A., Benito-Matías, L.F., Peñuelas-Rubira, J.L. & Suz*, L.M. (2017). Multi-cropping edible truffles and sweet chestnuts: production of high-quality Castanea sativa seedlings inoculated with Tuber aestivum, its ecotype T. uncinatum, T. brumale, and T. macrosporum. Mycorrhiza: DOI: 10.1007/s00572-017-0805-9. Available online. Ammirati, J.F., Niskanen*, T., Liimatainen*, K., Dimitar, B., Peintner, U., Kuhnert–Finkernagel, R. & Cripps, C. (2017). Spring and early summer species of Cortinarius, subgenus Telamonia, section Colymbadini and /Flavobasilis, in the mountains of western North America. Mycologia: DOI: 10.1080/00275514.2017.1349468. -

Drought-Tolerant Soybean Development: Evaluation of GM Lines Under Greenhouse and Field Conditions
JIRCAS Working Report No.91 57 Chapter 3-2 Drought-tolerant soybean development: evaluation of GM lines under greenhouse and field conditions Renata Fuganti-Pagliarini1,2, Silvana Regina Rockenbach Marin1,3, Mayla Daiane Côrrea Molinari3, Daniel de Amorim Barbosa3, Hugo Molinari4, Liliane Márcia Mertz- Henning1, Norman Neumaier1, José Renato Bouças Farias1, Kazuo Nakashima5, Kazuko Yamaguchi-Shinozaki6,7, Alexandre Lima Nepomuceno1 1 Embrapa Soja. Rodovia Carlos João Strass. Acesso Orlando Amaral. Warta. PO. Box 231 86001-970, Londrina, PR, Brazil 2 National Council for Scientific and Technological Development (CNPq), Brazil 3 Londrina State University – Biological Sciences Center. P.O. 10.011, 86057-970, Londrina, PR, Brazil 4 Embrapa Agroenergia. Parque Estação Biológica. s/nº, Brasília, Distrito Federal 70297-400, Brazil 5 Japan International Research Center for Agricultural Sciences, Tsukuba, Ibaraki 305-8686, Japan 6 Laboratory of Plant Molecular Physiology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo 113-8657, Japan 7 Research Institute for Agricultural and Life Sciences, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya-ku, Tokyo 156-8502, Japan * Corresponding author e-mail: [email protected] Abstract Drought is one of the greatest sources of environmental stress that has resulted in both economic and yield losses in many soybean-producing regions. The use of biotechnological tools aimed to produce plants with increased drought tolerance and productivity is the result of major investments in scientific and technological research. During the last decades, transcription factors (TFs) and key genes of important drought-responsive pathways have been used to develop genetically modified (GM) plants with increased tolerance to abiotic stress. -
A Molecular Phylogeny of Caraganeae (Leguminosae, Papilionoideae) Reveals Insights Into New Generic and Infrageneric Delimitations
A peer-reviewed open-access journal PhytoKeys 70: 111–137 (2016)Phylogeny of Caraganeae and new generic delimitations 111 doi: 10.3897/phytokeys.70.9641 RESEARCH ARTICLE http://phytokeys.pensoft.net Launched to accelerate biodiversity research A molecular phylogeny of Caraganeae (Leguminosae, Papilionoideae) reveals insights into new generic and infrageneric delimitations Lei Duan1, Xue Yang2, Peiliang Liu3, Gabriel Johnson4, Jun Wen4, Zhaoyang Chang3 1 Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, Guangdong 510650, P.R.China 2 Agriculture School, Kunming University, Kunming, Yunnan 650204, P.R.China 3 College of Life Sciences, Northwest A&F University, Yangling, Shaanxi 712100, China 4 Department of Botany, National Museum of Natural History, MRC 166, Smithsonian Institution, Washington DC, 20013-7012, U.S.A. Corresponding authors: Zhaoyang Chang ([email protected]); Jun Wen ([email protected]) Academic editor: Pavel Stoev | Received 22 June 2016 | Accepted 25 September 2016 | Published 4 October 2016 Citation: Duan L, Yang X, Liu P, Johnson G, Wen J, Chang Z (2016) A molecular phylogeny of Caraganeae (Leguminosae, Papilionoideae) reveals insights into new generic and infrageneric delimitations. PhytoKeys 70: 111–137. doi: 10.3897/ phytokeys.70.9641 Abstract Based on sequence data of nuclear ITS and plastid matK, trnL-F and psbA-trnH markers, the phylogeny of the subtribes Caraganinae and Chesneyinae in tribe Caraganeae was inferred. The results support the monophyly of each of the subtribes. Within subtribes Caraganinae, Calophaca and Halimodendron are herein transferred into Caragana to ensure its generic monophyly. The subtribe Chesneyinae is composed of four well-supported genera: Chesneya, Chesniella, Gueldenstaedtia and Tibetia.