A Focused Web Crawler Driven by Self-Optimizing Classifiers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Study of Web Crawler and Its Different Types
IOSR Journal of Computer Engineering (IOSR-JCE) e-ISSN: 2278-0661, p- ISSN: 2278-8727Volume 16, Issue 1, Ver. VI (Feb. 2014), PP 01-05 www.iosrjournals.org Study of Web Crawler and its Different Types Trupti V. Udapure1, Ravindra D. Kale2, Rajesh C. Dharmik3 1M.E. (Wireless Communication and Computing) student, CSE Department, G.H. Raisoni Institute of Engineering and Technology for Women, Nagpur, India 2Asst Prof., CSE Department, G.H. Raisoni Institute of Engineering and Technology for Women, Nagpur, India, 3Asso.Prof. & Head, IT Department, Yeshwantrao Chavan College of Engineering, Nagpur, India, Abstract : Due to the current size of the Web and its dynamic nature, building an efficient search mechanism is very important. A vast number of web pages are continually being added every day, and information is constantly changing. Search engines are used to extract valuable Information from the internet. Web crawlers are the principal part of search engine, is a computer program or software that browses the World Wide Web in a methodical, automated manner or in an orderly fashion. It is an essential method for collecting data on, and keeping in touch with the rapidly increasing Internet. This Paper briefly reviews the concepts of web crawler, its architecture and its various types. Keyword: Crawling techniques, Web Crawler, Search engine, WWW I. Introduction In modern life use of internet is growing in rapid way. The World Wide Web provides a vast source of information of almost all type. Now a day’s people use search engines every now and then, large volumes of data can be explored easily through search engines, to extract valuable information from web. -

Efficient Focused Web Crawling Approach for Search Engine
Ayar Pranav et al, International Journal of Computer Science and Mobile Computing, Vol.4 Issue.5, May- 2015, pg. 545-551 Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology ISSN 2320–088X IJCSMC, Vol. 4, Issue. 5, May 2015, pg.545 – 551 RESEARCH ARTICLE Efficient Focused Web Crawling Approach for Search Engine 1 2 Ayar Pranav , Sandip Chauhan Computer & Science Engineering, Kalol Institute of Technology and Research Canter, Kalol, Gujarat, India 1 [email protected]; 2 [email protected] Abstract— a focused crawler traverses the web, selecting out relevant pages to a predefined topic and neglecting those out of concern. Collecting domain specific documents using focused crawlers has been considered one of most important strategies to find relevant information. While surfing the internet, it is difficult to deal with irrelevant pages and to predict which links lead to quality pages. However most focused crawler use local search algorithm to traverse the web space, but they could easily trapped within limited a sub graph of the web that surrounds the starting URLs also there is problem related to relevant pages that are miss when no links from the starting URLs. There is some relevant pages are miss. To address this problem we design a focused crawler where calculating the frequency of the topic keyword also calculate the synonyms and sub synonyms of the keyword. The weight table is constructed according to the user query. To check the similarity of web pages with respect to topic keywords and priority of extracted link is calculated. -

Analysis and Evaluation of the Link and Content Based Focused Treasure-Crawler Ali Seyfi 1,2
Analysis and Evaluation of the Link and Content Based Focused Treasure-Crawler Ali Seyfi 1,2 1Department of Computer Science School of Engineering and Applied Sciences The George Washington University Washington, DC, USA. 2Centre of Software Technology and Management (SOFTAM) School of Computer Science Faculty of Information Science and Technology Universiti Kebangsaan Malaysia (UKM) Kuala Lumpur, Malaysia [email protected] Abstract— Indexing the Web is becoming a laborious task for search engines as the Web exponentially grows in size and distribution. Presently, the most effective known approach to overcome this problem is the use of focused crawlers. A focused crawler applies a proper algorithm in order to detect the pages on the Web that relate to its topic of interest. For this purpose we proposed a custom method that uses specific HTML elements of a page to predict the topical focus of all the pages that have an unvisited link within the current page. These recognized on-topic pages have to be sorted later based on their relevance to the main topic of the crawler for further actual downloads. In the Treasure-Crawler, we use a hierarchical structure called the T-Graph which is an exemplary guide to assign appropriate priority score to each unvisited link. These URLs will later be downloaded based on this priority. This paper outlines the architectural design and embodies the implementation, test results and performance evaluation of the Treasure-Crawler system. The Treasure-Crawler is evaluated in terms of information retrieval criteria such as recall and precision, both with values close to 0.5. Gaining such outcome asserts the significance of the proposed approach. -

Effective Focused Crawling Based on Content and Link Structure Analysis
(IJCSIS) International Journal of Computer Science and Information Security, Vol. 2, No. 1, June 2009 Effective Focused Crawling Based on Content and Link Structure Analysis Anshika Pal, Deepak Singh Tomar, S.C. Shrivastava Department of Computer Science & Engineering Maulana Azad National Institute of Technology Bhopal, India Emails: [email protected] , [email protected] , [email protected] Abstract — A focused crawler traverses the web selecting out dependent ontology, which in turn strengthens the metric used relevant pages to a predefined topic and neglecting those out of for prioritizing the URL queue. The Link-Structure-Based concern. While surfing the internet it is difficult to deal with method is analyzing the reference-information among the pages irrelevant pages and to predict which links lead to quality pages. to evaluate the page value. This kind of famous algorithms like In this paper, a technique of effective focused crawling is the Page Rank algorithm [5] and the HITS algorithm [6]. There implemented to improve the quality of web navigation. To check are some other experiments which measure the similarity of the similarity of web pages w.r.t. topic keywords, a similarity page contents with a specific subject using special metrics and function is used and the priorities of extracted out links are also reorder the downloaded URLs for the next crawl [7]. calculated based on meta data and resultant pages generated from focused crawler. The proposed work also uses a method for A major problem faced by the above focused crawlers is traversing the irrelevant pages that met during crawling to that it is frequently difficult to learn that some sets of off-topic improve the coverage of a specific topic. -
Report for Portal Specific
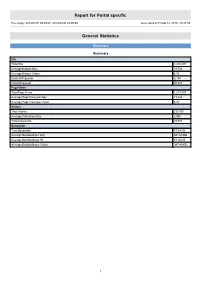
Report for Portal specific Time range: 2013/01/01 00:00:07 - 2013/03/31 23:59:59 Generated on Fri Apr 12, 2013 - 18:27:59 General Statistics Summary Summary Hits Total Hits 1,416,097 Average Hits per Day 15,734 Average Hits per Visitor 6.05 Cached Requests 8,154 Failed Requests 70,823 Page Views Total Page Views 1,217,537 Average Page Views per Day 13,528 Average Page Views per Visitor 5.21 Visitors Total Visitors 233,895 Average Visitors per Day 2,598 Total Unique IPs 49,753 Bandwidth Total Bandwidth 77.49 GB Average Bandwidth per Day 881.68 MB Average Bandwidth per Hit 57.38 KB Average Bandwidth per Visitor 347.40 KB 1 Activity Statistics Daily Daily Visitors 4,000 3,500 3,000 2,500 2,000 Visitors 1,500 1,000 500 0 2013/01/01 2013/01/15 2013/02/01 2013/02/15 2013/03/01 2013/03/15 Date Daily Hits 40,000 35,000 30,000 25,000 Hits 20,000 15,000 10,000 5,000 0 2013/01/01 2013/01/15 2013/02/01 2013/02/15 2013/03/01 2013/03/15 Date 2 Daily Bandwidth 1,300,000 1,200,000 1,100,000 1,000,000 900,000 800,000 700,000 600,000 500,000 Bandwidth (KB) 400,000 300,000 200,000 100,000 0 2013/01/01 2013/01/15 2013/02/01 2013/02/15 2013/03/01 2013/03/15 Date Daily Activity Date Hits Page Views Visitors Average Visit Length Bandwidth (KB) Sun 2013/02/10 11,783 10,245 2,280 13:01 648,207 Mon 2013/02/11 16,454 14,146 2,484 10:05 906,702 Tue 2013/02/12 19,572 17,089 3,062 07:47 926,190 Wed 2013/02/13 14,554 12,402 2,824 06:09 958,951 Thu 2013/02/14 12,577 10,666 2,690 05:03 821,129 Fri 2013/02/15 15,806 12,697 2,868 07:02 1,208,095 Sat 2013/02/16 16,811 14,939 -

The Google Search Engine
University of Business and Technology in Kosovo UBT Knowledge Center Theses and Dissertations Student Work Summer 6-2010 The Google search engine Ganimete Perçuku Follow this and additional works at: https://knowledgecenter.ubt-uni.net/etd Part of the Computer Sciences Commons Faculty of Computer Sciences and Engineering The Google search engine (Bachelor Degree) Ganimete Perçuku – Hasani June, 2010 Prishtinë Faculty of Computer Sciences and Engineering Bachelor Degree Academic Year 2008 – 2009 Student: Ganimete Perçuku – Hasani The Google search engine Supervisor: Dr. Bekim Gashi 09/06/2010 This thesis is submitted in partial fulfillment of the requirements for a Bachelor Degree Abstrakt Përgjithësisht makina kërkuese Google paraqitet si sistemi i kompjuterëve të projektuar për kërkimin e informatave në ueb. Google mundohet t’i kuptojë kërkesat e njerëzve në mënyrë “njerëzore”, dhe t’iu kthej atyre përgjigjen në formën të qartë. Por, ky synim nuk është as afër ideales dhe realizimi i tij sa vjen e vështirësohet me zgjerimin eksponencial që sot po përjeton ueb-i. Google, paraqitet duke ngërthyer në vetvete shqyrtimin e pjesëve që e përbëjnë, atyre në të cilat sistemi mbështetet, dhe rrethinave tjera që i mundësojnë sistemit të funksionojë pa probleme apo të përtërihet lehtë nga ndonjë dështim eventual. Procesi i grumbullimit të të dhënave ne Google dhe paraqitja e tyre në rezultatet e kërkimit ngërthen në vete regjistrimin e të dhënave nga ueb-faqe të ndryshme dhe vendosjen e tyre në rezervuarin e sistemit, përkatësisht në bazën e të dhënave ku edhe realizohen pyetësorët që kthejnë rezultatet e radhitura në mënyrën e caktuar nga algoritmi i Google. -

Scalability and Efficiency Challenges in Large-Scale Web Search
5/1/14 Scalability and Efficiency Challenges in Large-Scale Web Search Engines " Ricardo Baeza-Yates" B. Barla Cambazoglu! Yahoo Labs" Barcelona, Spain" Disclaimer Dis •# This talk presents the opinions of the authors. It does not necessarily reflect the views of Yahoo Inc. or any other entity." •# Algorithms, techniques, features, etc. mentioned here might or might not be in use by Yahoo or any other company." •# Some non-technical material (e.g., images) provided in this presentation were taken from the Web." 1 5/1/14 Yahoo Labs Barcelona •# Research topics" •# Web retrieval" –# web data mining" –# distributed web retrieval" –# semantic search" –# scalability and efficiency" –# social media" –# opinion/sentiment retrieval" –# web retrieval" –# personalization" Outline of the Tutorial •# Background (35 minutes)" •# Main sections" –# web crawling (75 minutes + 5 minutes Q/A)" –# indexing (75 minutes + 5 minutes Q/A)" –# query processing (90 minutes + 5 minutes Q/A)" –# caching (40 minutes + 5 minutes Q/A)" •# Concluding remarks (10 minutes)" •# Questions and open discussion (15 minutes)" 2 5/1/14 Structure of Main Sections •# Definitions" •# Metrics" •# Issues and techniques" –# single computer" –# cluster of computers" –# multiple search sites" •# Research problems" Background 3 5/1/14 Brief History of Search Engines •# Past" -# Before browsers" -# Gopher" -# Before the bubble" -# Altavista" -# Lycos" -# Infoseek" -# Excite" -# HotBot" •# Current" •# Future" -# After the bubble" •# Global" •# Facebook ?" -# Yahoo" •# Google, Bing" •# …" -

Finding OGC Web Services in the Digital Earth
Finding OGC Web Services in the Digital Earth Aneta J. Florczyk1, Patrick Mau´e2, Francisco J. L´opez-Pellicer1, and Javier Nogueras-Iso1 1 Departamento de Inform´atica e Ingenier´ıa de Sistemas, Universidad de Zaragoza, Spain {florczyk,fjlopez,jnog}@unizar.es 2 Institute for Geoinformatics (ifgi), University of Muenster, Germany [email protected] Abstract. The distribution of OGC Web Catalogues (CSW) across pro- fessional communities, the expert profile of the catalogue user and also the low coverage of OGC Web Services (OWS) in standard search engines reduce the possibility of discovery and sharing geographic information. This paper presents an approach to simple spatio-temporal search of OWS retrieved from the web by a specialized crawler. The Digital Earth could benefit from this solution as it overcomes technical boundaries and solves the limitation of CSW acceptance only within professional com- munity. Keywords: OWS, discovery, open search 1 Introduction The concept of the Digital Earth envisions easy-to-use applications to support non-expert users to interact with various kinds of geographic information. Users are expected to search, load, and visualize spatial data without being challenged by the plethora of file formats and methods to access and download the data. The Open Geospatial Consortium (OGC) standards specify well-defined interfaces for spatial data services to ensure interoperability across information communities. Embedded in Spatial Data Infrastructures (SDI), OGC services are mainly (but not only) used by the public administration to publish their data on the Web. On-going initiatives such as the Global Earth Observation System of Systems (GEOSS) or the European Shared Environmental Information Space (SEIS) form an important backbone to the vision of the Digital Earth. -

An Intelligent Meta Search Engine for Efficient Web Document Retrieval
IOSR Journal of Computer Engineering (IOSR-JCE) e-ISSN: 2278-0661,p-ISSN: 2278-8727, Volume 17, Issue 2, Ver. V (Mar – Apr. 2015), PP 45-54 www.iosrjournals.org An Intelligent Meta Search Engine for Efficient Web Document Retrieval 1 2 Keerthana.I.P , Aby Abahai.T 1(Dept. of CSE, Mar Athanasius College of Engineering, Kothamangalam , Kerala, India) 2(Dept. of CSE, Assistant Professor, Mar Athanasius College of Engineering, Kothamangalam, Kerala, India) Abstract: In daily use of internet, when searching information we face lots of difficulty due to the rapid growth of Information Resources. This is because of the fact that a single search engine cannot index the entire web of resources. A Meta Search Engine is a solution for this, which submits the query to many other search engines and returns summary of the results. Therefore, the search results receive are an aggregate result of multiple searches. This strategy gives our search a boarder scope than searching a single search engine, but the results are not always better. Because the Meta Search Engine must use its own algorithm to choose the best results from multiple search engines. In this paper we proposed a new Meta Search Engine to overcome these drawbacks. It uses a new page rank algorithm called modified ranking for ranking and optimizing the search results in an efficient way. It is a two phase ranking algorithm used for ordering the web pages based on their relevance and popularity. This Meta Search Engine is developed in such a way that it will produce more efficient results than traditional search engines. -

Collecte Orientée Sur Le Web Pour La Recherche D'information Spécialisée
Collecte orientée sur le Web pour la recherche d’information spécialisée Clément de Groc To cite this version: Clément de Groc. Collecte orientée sur le Web pour la recherche d’information spécialisée. Autre [cs.OH]. Université Paris Sud - Paris XI, 2013. Français. NNT : 2013PA112073. tel-00853250 HAL Id: tel-00853250 https://tel.archives-ouvertes.fr/tel-00853250 Submitted on 22 Aug 2013 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Université Paris-Sud École doctorale d’informatique LIMSI-CNRS Collecte orientée sur le Web pour la recherche d’information spécialisée par Clément de Groc Thèse présentée pour obtenir le grade de Docteur de l’Université Paris-Sud Soutenue publiquement à Orsay le mercredi 5 juin 2013 en présence d’un jury composé de : Rapporteur Éric Gaussier Université de Grenoble Rapporteur Jacques Savoy Université de Neuchâtel Examinatrice Chantal Reynaud Université Paris-Sud Examinateur Mohand Boughanem Université de Toulouse Directeur Pierre Zweigenbaum CNRS Co-directeur Xavier Tannier Université Paris-Sud Invité Claude de Loupy Syllabs REMERCIEMENTS Cette thèse est l’aboutissement d’un travail individuel qui a nécessité le soutien et la participation de nombreuses personnes. Je remercie messieurs Éric Gaussier et Jacques Savoy de m’avoir fait l’immense honneur d’accep- ter d’être rapporteurs de mon travail, au même titre que madame Chantal Reynaud et monsieur Mohand Boughanem qui ont accepté de faire partie de mon jury. -

Web Crawling with Carlos Castillo
Modern Information Retrieval Chapter 12 Web Crawling with Carlos Castillo Applications of a Web Crawler Architecture and Implementation Scheduling Algorithms Crawling Evaluation Extensions Examples of Web Crawlers Trends and Research Issues Web Crawling, Modern Information Retrieval, Addison Wesley, 2010 – p. 1 Introduction and a Brief History A Web Crawler is a software for downloading pages from the Web Also known as Web Spider, Web Robot, or simply Bot Cycle of a Web crawling process The crawler start downloading a set of seed pages, that are parsed and scanned for new links The links to pages that have not yet been downloaded are added to a central queue for download later Next, the crawler selects a new page for download and the process is repeated until a stop criterion is met Web Crawling, Modern Information Retrieval, Addison Wesley, 2010 – p. 2 Introduction and a Brief History The first known web crawler was created in 1993 by Matthew Gray, an undergraduate student at MIT On June of that year, Gray sent the following message to the www-talk mailing list: “I have written a perl script that wanders the WWW collecting URLs, keeping tracking of where it’s been and new hosts that it finds. Eventually, after hacking up the code to return some slightly more useful information (currently it just returns URLs), I will produce a searchable index of this.” The project of this Web crawler was called WWWW (World Wide Web Wanderer) It was used mostly for Web characterization studies Web Crawling, Modern Information Retrieval, Addison Wesley, 2010 – p. 3 Introduction and a Brief History On June 1994, Brian Pinkerton, a PhD student at the University of Washington, posted the following message to the comp.infosystems.announce news “The WebCrawler Index is now available for searching! The index is broad: it contains information from as many different servers as possible. -

Using Web Crawler Technology for Text Analysis Ofgeo-Events: a Case Study of the Huangyan Island Incident
The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Volume XL-4/W3, 2013 ISPRS/IGU/ICA Joint Workshop on Borderlands Modelling and Understanding for Global Sustainability 2013, 5 – 6 December 2013, Beijing, China USING WEB CRAWLER TECHNOLOGY FOR TEXT ANALYSIS OFGEO-EVENTS: A CASE STUDY OF THE HUANGYAN ISLAND INCIDENT HU. Hao, GE. Yuejing School of Geography Beijing Normal University 100875 No. 19 Xin Jie Kou Wai Street, Haidian District, Beijing, P.R. China - [email protected] [email protected] Commission KEY WORDS: web crawler technology; text information; sentiment analysis; Huangyan Island incident ABSTRACT: With the social networking and network socialisation have brought more text information and social relationships into our daily lives, the question of whether big data can be fully used to study the phenomenon and discipline of natural sciences has prompted many specialists and scholars to innovate their research. Though politics were integrally involved in the hyperlinked word issues since 1990s, automatic assembly of different geospatial web and distributed geospatial information systems utilizing service chaining have explored and built recently, the information collection and data visualisation of geo-events have always faced the bottleneck of traditional manual analysis because of the sensibility, complexity, relativity, timeliness and unexpected characteristics of political events. Based on the framework of Heritrix and the analysis of web-based text, word frequency, sentiment tendency and dissemination path of the Huangyan Island incident is studied here by combining web crawler technology and the text analysis method. The results indicate that tag cloud, frequency map, attitudes pie, individual mention ratios and dissemination flow graph based on the data collection and processing not only highlight the subject and theme vocabularies of related topics but also certain issues and problems behind it.