Y-Aminobutyric Acid Uptake by a Bacterial System With
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Review of Market for Octane Enhancers
May 2000 • NREL/SR-580-28193 Review of Market for Octane Enhancers Final Report J.E. Sinor Consultants, Inc. Niwot, Colorado National Renewable Energy Laboratory 1617 Cole Boulevard Golden, Colorado 80401-3393 NREL is a U.S. Department of Energy Laboratory Operated by Midwest Research Institute • Battelle • Bechtel Contract No. DE-AC36-99-GO10337 May 2000 • NREL/SR-580-28193 Review of Market for Octane Enhancers Final Report J.E. Sinor Consultants, Inc. Niwot, Colorado NREL Technical Monitor: K. Ibsen Prepared under Subcontract No. TXE-0-29113-01 National Renewable Energy Laboratory 1617 Cole Boulevard Golden, Colorado 80401-3393 NREL is a U.S. Department of Energy Laboratory Operated by Midwest Research Institute • Battelle • Bechtel Contract No. DE-AC36-99-GO10337 NOTICE This report was prepared as an account of work sponsored by an agency of the United States government. Neither the United States government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States government or any agency thereof. Available electronically at http://www.doe.gov/bridge Available for a processing fee to U.S. -

Precursors and Chemicals Frequently Used in the Illicit Manufacture of Narcotic Drugs and Psychotropic Substances 2017
INTERNATIONAL NARCOTICS CONTROL BOARD Precursors and chemicals frequently used in the illicit manufacture of narcotic drugs and psychotropic substances 2017 EMBARGO Observe release date: Not to be published or broadcast before Thursday, 1 March 2018, at 1100 hours (CET) UNITED NATIONS CAUTION Reports published by the International Narcotics Control Board in 2017 The Report of the International Narcotics Control Board for 2017 (E/INCB/2017/1) is supplemented by the following reports: Narcotic Drugs: Estimated World Requirements for 2018—Statistics for 2016 (E/INCB/2017/2) Psychotropic Substances: Statistics for 2016—Assessments of Annual Medical and Scientific Requirements for Substances in Schedules II, III and IV of the Convention on Psychotropic Substances of 1971 (E/INCB/2017/3) Precursors and Chemicals Frequently Used in the Illicit Manufacture of Narcotic Drugs and Psychotropic Substances: Report of the International Narcotics Control Board for 2017 on the Implementation of Article 12 of the United Nations Convention against Illicit Traffic in Narcotic Drugs and Psychotropic Substances of 1988 (E/INCB/2017/4) The updated lists of substances under international control, comprising narcotic drugs, psychotropic substances and substances frequently used in the illicit manufacture of narcotic drugs and psychotropic substances, are contained in the latest editions of the annexes to the statistical forms (“Yellow List”, “Green List” and “Red List”), which are also issued by the Board. Contacting the International Narcotics Control Board The secretariat of the Board may be reached at the following address: Vienna International Centre Room E-1339 P.O. Box 500 1400 Vienna Austria In addition, the following may be used to contact the secretariat: Telephone: (+43-1) 26060 Fax: (+43-1) 26060-5867 or 26060-5868 Email: [email protected] The text of the present report is also available on the website of the Board (www.incb.org). -

Organocatalytic Asymmetric N-Sulfonyl Amide C-N Bond Activation to Access Axially Chiral Biaryl Amino Acids
ARTICLE https://doi.org/10.1038/s41467-020-14799-8 OPEN Organocatalytic asymmetric N-sulfonyl amide C-N bond activation to access axially chiral biaryl amino acids Guanjie Wang1, Qianqian Shi2, Wanyao Hu1, Tao Chen1, Yingying Guo1, Zhouli Hu1, Minghua Gong1, ✉ ✉ ✉ Jingcheng Guo1, Donghui Wei 2 , Zhenqian Fu 1,3 & Wei Huang1,3 1234567890():,; Amides are among the most fundamental functional groups and essential structural units, widely used in chemistry, biochemistry and material science. Amide synthesis and trans- formations is a topic of continuous interest in organic chemistry. However, direct catalytic asymmetric activation of amide C-N bonds still remains a long-standing challenge due to high stability of amide linkages. Herein, we describe an organocatalytic asymmetric amide C-N bonds cleavage of N-sulfonyl biaryl lactams under mild conditions, developing a general and practical method for atroposelective construction of axially chiral biaryl amino acids. A structurally diverse set of axially chiral biaryl amino acids are obtained in high yields with excellent enantioselectivities. Moreover, a variety of axially chiral unsymmetrical biaryl organocatalysts are efficiently constructed from the resulting axially chiral biaryl amino acids by our present strategy, and show competitive outcomes in asymmetric reactions. 1 Key Laboratory of Flexible Electronics & Institute of Advanced Materials, Jiangsu National Synergetic Innovation Center for Advanced Materials, Nanjing Tech University, 30 South Puzhu Road, Nanjing 211816, China. 2 College -

GABA Receptors
D Reviews • BIOTREND Reviews • BIOTREND Reviews • BIOTREND Reviews • BIOTREND Reviews Review No.7 / 1-2011 GABA receptors Wolfgang Froestl , CNS & Chemistry Expert, AC Immune SA, PSE Building B - EPFL, CH-1015 Lausanne, Phone: +41 21 693 91 43, FAX: +41 21 693 91 20, E-mail: [email protected] GABA Activation of the GABA A receptor leads to an influx of chloride GABA ( -aminobutyric acid; Figure 1) is the most important and ions and to a hyperpolarization of the membrane. 16 subunits with γ most abundant inhibitory neurotransmitter in the mammalian molecular weights between 50 and 65 kD have been identified brain 1,2 , where it was first discovered in 1950 3-5 . It is a small achiral so far, 6 subunits, 3 subunits, 3 subunits, and the , , α β γ δ ε θ molecule with molecular weight of 103 g/mol and high water solu - and subunits 8,9 . π bility. At 25°C one gram of water can dissolve 1.3 grams of GABA. 2 Such a hydrophilic molecule (log P = -2.13, PSA = 63.3 Å ) cannot In the meantime all GABA A receptor binding sites have been eluci - cross the blood brain barrier. It is produced in the brain by decarb- dated in great detail. The GABA site is located at the interface oxylation of L-glutamic acid by the enzyme glutamic acid decarb- between and subunits. Benzodiazepines interact with subunit α β oxylase (GAD, EC 4.1.1.15). It is a neutral amino acid with pK = combinations ( ) ( ) , which is the most abundant combi - 1 α1 2 β2 2 γ2 4.23 and pK = 10.43. -
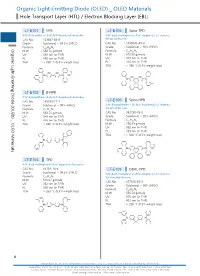
Organic Light-Emitting Diode (OLED) OLED Materials Hole Transport Layer (HTL) / Electron Blocking Layer (EBL)
Organic Light-EmittingOrganic Light-Emitting DiodeHole Transport(OLED) Layer (HTL) / Diode (OLED) _ OLED Materials Electron Blocking Layer (EBL) Organic Light-Emitting Diode (OLED) _ OLED Materials Hole Transport Layer (HTL) / Electron Blocking Layer (EBL) LT-E101 NPB LT-E105 Spiro-TPD N,N'-Bis(naphthalen-1-yl)-N,N'-bis(phenyl)-benzidine N,N'-Bis(3-methylphenyl)-N,N'-bis(phenyl)-2,7-diamino- CAS No. : 123847-85-8 9,9-spirobifluorene Grade : Sublimed, > 99.5% (HPLC) CAS No. : 1033035-83-4 : Grade : Sublimed, > 99% (HPLC) Organic Light-Emitting Diode (OLED) _ OLED Materials Formula C44H32N2 M.W. : 588.74 g/mole Formula : C51H38N2 UV : 339 nm (in THF) M.W. : 678.86 g/mole PL : 450 nm (in THF) UV : 379 nm (in THF) TGA : > 350 °C (0.5% weight loss) PL : 416 nm (in THF) TGA : > 280 °C (0.5% weight loss) N N N N LT-E102 β-NPB N,N'-Bis(naphthalen-2-yl)-N,N'-bis(phenyl)-benzidine Spiro-NPB CAS No. : 139255-17-7 LT-E106 Grade : Sublimed, > 99% (HPLC) N,N'-Bis(naphthalen-1-yl)-N,N'-bis(phenyl)-2,7-diamino- 9,9-spirobifluorene Formula : C44H32N2 M.W. : 588.74 g/mole CAS No. : 932739-76-9 UV : 349 nm (in THF) Grade : Sublimed, > 99% (HPLC) PL : 416 nm (in THF) Formula : C57H38N2 TGA : > 330 °C (0.5% weight loss) M.W. : 750.93 g/mole UV : 380 nm (in THF) PL : 453 nm (in THF) TGA : > 360 °C (0.5% weight loss) N N N N LT-E103 TPD N,N'-Bis(3-methylphenyl)-N,N'-bis(phenyl)-benzidine CAS No. -

Endogenous Metabolites: JHU NIMH Center Page 1
S. No. Amino Acids (AA) 24 L-Homocysteic acid 1 Glutaric acid 25 L-Kynurenine 2 Glycine 26 N-Acetyl-Aspartic acid 3 L-arginine 27 N-Acetyl-L-alanine 4 L-Aspartic acid 28 N-Acetyl-L-phenylalanine 5 L-Glutamine 29 N-Acetylneuraminic acid 6 L-Histidine 30 N-Methyl-L-lysine 7 L-Isoleucine 31 N-Methyl-L-proline 8 L-Leucine 32 NN-Dimethyl Arginine 9 L-Lysine 33 Norepinephrine 10 L-Methionine 34 Phenylacetyl-L-glutamine 11 L-Phenylalanine 35 Pyroglutamic acid 12 L-Proline 36 Sarcosine 13 L-Serine 37 Serotonin 14 L-Tryptophan 38 Stachydrine 15 L-Tyrosine 39 Taurine 40 Urea S. No. AA Metabolites and Conjugates 1 1-Methyl-L-histidine S. No. Carnitine conjugates 2 2-Methyl-N-(4-Methylphenyl)alanine 1 Acetyl-L-carnitine 3 3-Methylindole 2 Butyrylcarnitine 4 3-Methyl-L-histidine 3 Decanoyl-L-carnitine 5 4-Aminohippuric acid 4 Isovalerylcarnitine 6 5-Hydroxylysine 5 Lauroyl-L-carnitine 7 5-Hydroxymethyluracil 6 L-Glutarylcarnitine 8 Alpha-Aspartyl-lysine 7 Linoleoylcarnitine 9 Argininosuccinic acid 8 L-Propionylcarnitine 10 Betaine 9 Myristoyl-L-carnitine 11 Betonicine 10 Octanoylcarnitine 12 Carnitine 11 Oleoyl-L-carnitine 13 Creatine 12 Palmitoyl-L-carnitine 14 Creatinine 13 Stearoyl-L-carnitine 15 Dimethylglycine 16 Dopamine S. No. Krebs Cycle 17 Epinephrine 1 Aconitate 18 Hippuric acid 2 Citrate 19 Homo-L-arginine 3 Ketoglutarate 20 Hydroxykynurenine 4 Malate 21 Indolelactic acid 5 Oxalo acetate 22 L-Alloisoleucine 6 Succinate 23 L-Citrulline 24 L-Cysteine-glutathione disulfide Semi-quantitative analysis of endogenous metabolites: JHU NIMH Center Page 1 25 L-Glutathione, reduced Table 1: Semi-quantitative analysis of endogenous molecules and their derivatives by Liquid Chromatography- Mass Spectrometry (LC-TripleTOF “or” LC-QTRAP). -

Opposing Effects of Dehydroepiandrosterone And
European Journal of Endocrinology (2000) 143 687±695 ISSN 0804-4643 EXPERIMENTAL STUDY Opposing effects of dehydroepiandrosterone and dexamethasone on the generation of monocyte-derived dendritic cells M O Canning, K Grotenhuis, H J de Wit and H A Drexhage Department of Immunology, Erasmus University Rotterdam, The Netherlands (Correspondence should be addressed to H A Drexhage, Lab Ee 838, Department of Immunology, Erasmus University, PO Box 1738, 3000 DR Rotterdam, The Netherlands; Email: [email protected]) Abstract Background: Dehydroepiandrosterone (DHEA) has been suggested as an immunostimulating steroid hormone, of which the effects on the development of dendritic cells (DC) are unknown. The effects of DHEA often oppose those of the other adrenal glucocorticoid, cortisol. Glucocorticoids (GC) are known to suppress the immune response at different levels and have recently been shown to modulate the development of DC, thereby influencing the initiation of the immune response. Variations in the duration of exposure to, and doses of, GC (particularly dexamethasone (DEX)) however, have resulted in conflicting effects on DC development. Aim: In this study, we describe the effects of a continuous high level of exposure to the adrenal steroid DHEA (1026 M) on the generation of immature DC from monocytes, as well as the effects of the opposing steroid DEX on this development. Results: The continuous presence of DHEA (1026 M) in GM-CSF/IL-4-induced monocyte-derived DC cultures resulted in immature DC with a morphology and functional capabilities similar to those of typical immature DC (T cell stimulation, IL-12/IL-10 production), but with a slightly altered phenotype of increased CD80 and decreased CD43 expression (markers of maturity). -

Fluorinated Analogues of Glutamic Acid and Glutamine R
γ-Fluorinated analogues of glutamic acid and glutamine R. Dave, B. Badet, Patrick Meffre To cite this version: R. Dave, B. Badet, Patrick Meffre. γ-Fluorinated analogues of glutamic acid and glutamine. Amino Acids, Springer Verlag, 2003, 24 (3), pp.245-261. 10.1007/s00726-002-0410-9. hal-02002676 HAL Id: hal-02002676 https://hal.archives-ouvertes.fr/hal-02002676 Submitted on 11 Feb 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. γ-Fluorinated analogues of glutamic acid and glutamine Review Article 1 2 1 R. Dave , B. Badet , and P. Meffre 1 UMR 7573-C.N.R.S., ENSCP, Paris, France 2 UPR 2301-CNRS, ICSN, Gif-sur-Yvette, France Summary. γ-Fluorinated analogues of glutamic acid and glutamine N-bromosuccinimide; NFSi, N-fluorobenzenesulfonimide; NMR, are compounds of biological interest. Syntheses of such compounds nuclear magnetic resonance; 2-PrOH, isopropanol; PTSA, p- are extensively reviewed in this article. 4-Fluoroglutamic acid toluenesulfonic acid; TCDI, thiocarbonyldiimidazole; TEMPO, was prepared as a mixture of racemic diastereomers by Michael 2,2,6,6-tetramethyl piperidine-1-oxyl; TFA, trifluoroacetic acid. reaction, inverse-Michael reaction or by electrophilic / nucleophilic fluorination. -

NINDS Custom Collection II
ACACETIN ACEBUTOLOL HYDROCHLORIDE ACECLIDINE HYDROCHLORIDE ACEMETACIN ACETAMINOPHEN ACETAMINOSALOL ACETANILIDE ACETARSOL ACETAZOLAMIDE ACETOHYDROXAMIC ACID ACETRIAZOIC ACID ACETYL TYROSINE ETHYL ESTER ACETYLCARNITINE ACETYLCHOLINE ACETYLCYSTEINE ACETYLGLUCOSAMINE ACETYLGLUTAMIC ACID ACETYL-L-LEUCINE ACETYLPHENYLALANINE ACETYLSEROTONIN ACETYLTRYPTOPHAN ACEXAMIC ACID ACIVICIN ACLACINOMYCIN A1 ACONITINE ACRIFLAVINIUM HYDROCHLORIDE ACRISORCIN ACTINONIN ACYCLOVIR ADENOSINE PHOSPHATE ADENOSINE ADRENALINE BITARTRATE AESCULIN AJMALINE AKLAVINE HYDROCHLORIDE ALANYL-dl-LEUCINE ALANYL-dl-PHENYLALANINE ALAPROCLATE ALBENDAZOLE ALBUTEROL ALEXIDINE HYDROCHLORIDE ALLANTOIN ALLOPURINOL ALMOTRIPTAN ALOIN ALPRENOLOL ALTRETAMINE ALVERINE CITRATE AMANTADINE HYDROCHLORIDE AMBROXOL HYDROCHLORIDE AMCINONIDE AMIKACIN SULFATE AMILORIDE HYDROCHLORIDE 3-AMINOBENZAMIDE gamma-AMINOBUTYRIC ACID AMINOCAPROIC ACID N- (2-AMINOETHYL)-4-CHLOROBENZAMIDE (RO-16-6491) AMINOGLUTETHIMIDE AMINOHIPPURIC ACID AMINOHYDROXYBUTYRIC ACID AMINOLEVULINIC ACID HYDROCHLORIDE AMINOPHENAZONE 3-AMINOPROPANESULPHONIC ACID AMINOPYRIDINE 9-AMINO-1,2,3,4-TETRAHYDROACRIDINE HYDROCHLORIDE AMINOTHIAZOLE AMIODARONE HYDROCHLORIDE AMIPRILOSE AMITRIPTYLINE HYDROCHLORIDE AMLODIPINE BESYLATE AMODIAQUINE DIHYDROCHLORIDE AMOXEPINE AMOXICILLIN AMPICILLIN SODIUM AMPROLIUM AMRINONE AMYGDALIN ANABASAMINE HYDROCHLORIDE ANABASINE HYDROCHLORIDE ANCITABINE HYDROCHLORIDE ANDROSTERONE SODIUM SULFATE ANIRACETAM ANISINDIONE ANISODAMINE ANISOMYCIN ANTAZOLINE PHOSPHATE ANTHRALIN ANTIMYCIN A (A1 shown) ANTIPYRINE APHYLLIC -

Hydroxydopamine Lesions of the Nucleus Accumbens and the Mglur2/3 Agonist LY379268
Neuropsychopharmacology (2003) 28, 1440–1447 & 2003 Nature Publishing Group All rights reserved 0893-133X/03 $25.00 www.neuropsychopharmacology.org Toluene-Induced Locomotor Activity is Blocked by 6- Hydroxydopamine Lesions of the Nucleus Accumbens and the mGluR2/3 Agonist LY379268 1,3 2 ,1 AC Riegel , SF Ali , ED French* 1Department of Pharmacology, College of Medicine, University of Arizona, Tucson, AZ, USA; 2Neurochemistry Laboratory, Division of Neurotoxicology, National Center for Toxicological Research/US FDA, Jefferson, AR, USA The abuse of volatile inhalants remains a prominent, yet poorly understood, form of substance abuse among youth. Nevertheless, the identification of a mechanism underlying the reinforcing properties of inhalants has been hampered by the lack of a clearly identifiable neural substrate upon which these chemicals act. One ingredient that is common to many abused inhalants is toluene, an organic solvent that is self-administered by nonhuman primates and rodents. Most drugs of abuse have been found to elicit forward locomotion in rats, an effect owing to the activation of mesoaccumbal dopamine (DA) pathways. Thus, the present study was undertaken using two different approaches to determine whether toluene-induced locomotor hyperactivity is also ultimately dependent upon DA neurotransmission in the mesolimbic nucleus accumbens (NAC). Here we report on the effects of 6-hydroxydopamine (6-OHDA) lesions of the NAC or pretreatment with the metabotropic mGlu2/3 receptor agonist LY379268 on toluene-induced locomotor activity. Both procedures, which are known to alter neurotransmission within the NAC, significantly attenuated toluene’s locomotor stimulatory effects. These results provide strong support for a central mechanism of action of inhalants, which in the past has been more typically attributed to general nonspecific mechanisms throughout the brain. -

Inhalant Abuse Pediatric Care
CLINICAL REPORT Guidance for the Clinician in Rendering Inhalant Abuse Pediatric Care Janet F. Williams, MD, Michael Storck, MD, and the Committee on Substance Abuse and Committee on Native American Child Health ABSTRACT Inhalant abuse is the intentional inhalation of a volatile substance for the purpose of achieving an altered mental state. As an important, yet-underrecognized form of substance abuse, inhalant abuse crosses all demographic, ethnic, and socioeco- nomic boundaries, causing significant morbidity and mortality in school-aged and older children. This clinical report reviews key aspects of inhalant abuse, empha- sizes the need for greater awareness, and offers advice regarding the pediatrician’s role in the prevention and management of this substance abuse problem. TYPES OF CHEMICALS AND PRODUCTS ABUSED The term “inhalant” encompasses a wide range of pharmacologically diverse substances that readily vaporize. Most other substances of abuse are classified by grouping together substances that share a specific central nervous system action or perceived psychoactive effect, but inhalant substances that are abused are grouped by having a common route of drug use. Inhalant abuse, sometimes referred to as solvent or volatile substance abuse, can be better understood when the expansive list of inhalants is classified into 3 groups on the basis of what is currently known pharmacologically: group I includes volatile solvents, fuels, and anesthetics; group II includes nitrous oxide; and group III includes volatile alkyl nitrites (Table 1). This classification is also consistent with reported differences in user populations, patterns of abuse, and associated problems seen clinically.1–3 Drugs that do not www.pediatrics.org/cgi/doi/10.1542/ readily vaporize at room temperature, such as cocaine, heroin, nicotine, or alcohol, peds.2007-0470 can also be abused through inhalation, but characteristic pharmacologic properties doi:10.1542/peds.2007-0470 distinguish these substances from inhalants. -

The Role of Glutamine in the Complex Interaction Between Gut Microbiota and Health: a Narrative Review
International Journal of Molecular Sciences Review The Role of Glutamine in the Complex Interaction between Gut Microbiota and Health: A Narrative Review 1, , 1 1, 1 Simone Perna * y , Tariq A. Alalwan , Zahraa Alaali y , Tahera Alnashaba , Clara Gasparri 2, Vittoria Infantino 3 , Layla Hammad 1, Antonella Riva 4, Giovanna Petrangolini 4 , Pietro Allegrini 4 and Mariangela Rondanelli 5,6 1 Department of Biology, College of Science, University of Bahrain, 32038 Sakhir, Bahrain; [email protected] (T.A.A.); [email protected] (Z.A.); [email protected] (T.A.); [email protected] (L.H.) 2 Endocrinology and Nutrition Unit, Azienda di Servizi alla Persona “Istituto Santa Margherita”, University of Pavia, Pavia 27100, Italy; [email protected] 3 Department of Biomedical Science and Human Oncology, University of Bari, Bari 70121, Italy; [email protected] 4 Research and Development Department, Indena SpA, 20139 Milan, Italy; [email protected] (A.R.); [email protected] (G.P.); [email protected] (P.A.) 5 IRCCS Mondino Foundation, Pavia 27100, Italy; [email protected] 6 Department of Public Health, Experimental and Forensic Medicine, Unit of Human and Clinical Nutrition, University of Pavia, Pavia 27100, Italy * Correspondence: [email protected] or [email protected]; Tel.: +973-39-37-99-46 The two authors contributed equally to this work. y Received: 21 August 2019; Accepted: 20 October 2019; Published: 22 October 2019 Abstract: The scientific literature has demonstrated that glutamine is one of the main beneficial amino acids. It plays an important role in gut microbiota and immunity. This paper provides a critical overview of experimental studies (in vitro, in vivo, and clinical) investigating the efficacy of glutamine and its effect on gut microbiota.