Supplementary Table S1. Summary of the Six Next-Generation Sequencing (NGS) Studies Containing 241 Paired Melanoma Tumor/Normal Samples
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Germline Mutations Affecting Gα11 in Hypoparathyroidism
T h e new england journal o f medicine of a meta-analysis of studies of the effect of re- terone levels were increased. Although long- duced dietary salt on the incidence of cardiovas- term, modest reductions in salt intake result in cular events and death.1 The authors of the Co- small, physiologic increases in plasma renin ac- chrane report wrote that there was “no strong tivity,3 the preponderance of evidence suggests evidence of benefit.” In a summary statement, we that a reduced salt intake is associated with a wrote that this particular Cochrane analysis con- decreased risk of cardiovascular events and cluded that reducing dietary salt intake did not death. Furthermore, it is worth remembering decrease the risk of death or cardiovascular dis- that diuretics remain one of the most effective ease. Stigler et al. suggest that “indeterminate antihypertensive therapies, and their beneficial results,” rather than no significant effect, would effect on cardiovascular disease is well docu- be a more appropriate interpretation of the analy- mented.4 Nevertheless, as we suggested, in sis. Both interpretations may be correct. Although terms of safety, the lower limit of salt consump- it may not be possible to reject the null hypothe- tion has not been clearly defined. sis with certainty (i.e., no effect of reduced salt), Theodore A. Kotchen, M.D. the analysis should have been powered to detect Allen W. Cowley, Jr., Ph.D. a clinically meaningful difference, and the con- Medical College of Wisconsin clusion of no effect provides more guidance than Milwaukee, WI “indeterminate results” to clinical decision mak- [email protected] ers. -

ACVR1 Antibody Cat
ACVR1 Antibody Cat. No.: 4791 Western blot analysis of ACVR1 in A549 cell lysate with ACVR1 antibody at 1 μg/mL in (A) the absence and (B) the presence of blocking peptide. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human, Mouse HOMOLOGY: Predicted species reactivity based on immunogen sequence: Bovine: (100%), Rat: (93%) ACVR1 antibody was raised against a 14 amino acid synthetic peptide near the amino terminus of the human ACVR1. IMMUNOGEN: The immunogen is located within the first 50 amino acids of ACVR1. TESTED APPLICATIONS: ELISA, WB ACVR1 antibody can be used for detection of ACVR1 by Western blot at 1 μg/mL. APPLICATIONS: Antibody validated: Western Blot in human samples. All other applications and species not yet tested. At least four isoforms of ACVR1 are known to exist. This antibody is predicted to have no SPECIFICITY: cross-reactivity to ACVR1B or ACVR1C. POSITIVE CONTROL: 1) Cat. No. 1203 - A549 Cell Lysate Properties October 1, 2021 1 https://www.prosci-inc.com/acvr1-antibody-4791.html PURIFICATION: ACVR1 Antibody is affinity chromatography purified via peptide column. CLONALITY: Polyclonal ISOTYPE: IgG CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: ACVR1 Antibody is supplied in PBS containing 0.02% sodium azide. CONCENTRATION: 1 mg/mL ACVR1 antibody can be stored at 4˚C for three months and -20˚C, stable for up to one STORAGE CONDITIONS: year. As with all antibodies care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: ACVR1 ACVR1 Antibody: FOP, ALK2, SKR1, TSRI, ACTRI, ACVR1A, ACVRLK2, Activin receptor type-1, ALTERNATE NAMES: Activin receptor type I, ACTR-I ACCESSION NO.: NP_001096 PROTEIN GI NO.: 4501895 GENE ID: 90 USER NOTE: Optimal dilutions for each application to be determined by the researcher. -

'Stress–Response' Kinase Pathways in Alzheimer's Disease Progression
Available online at www.sciencedirect.com ScienceDirect Involvement of ‘stress–response’ kinase pathways in Alzheimer’s disease progression 1 Georges Mairet-Coello and Franck Polleux Alzheimer’s disease (AD) is the most prevalent cause of oligomers of Ab are causal to synaptic toxicity [2], trigger dementia, affecting more than 25 million people worldwide. synaptic dysfunction, synapse loss and impaired long- Current models of the pathophysiological mechanisms of AD term potentiation (LTP) [3]. The exact nature of the suggest that the accumulation of soluble oligomeric forms of Ab species (dimers, trimers, Ab*56, protofibrils) respon- amyloid-b (Ab) peptides causes early loss of excitatory sible for this early synaptotoxicity is still under investi- synapses and impairs synaptic plasticity. The signaling gation [4]. Most experimental designs use a mixture of pathways mediating Ab oligomer-induced impairment of various forms of synthetic Ab oligomers, or natural Ab synaptic plasticity and loss of excitatory synapses are only oligomers isolated from the brain of AD subjects, to beginning to be unraveled. Here, we review recent evidence induce rapid loss of excitatory synapses in hippocampal supporting the critical contribution of conserved ‘stress– and cortical neurons in vitro [5–10]. Transgenic mouse response’ kinase pathways in AD progression. models of AD engineered to overexpress human mutant forms of Amyloid Precursor Protein (hAPP), with or with- Addresses out overexpression of mutant presenilins (PS), produce The Scripps Research Institute, Dorris Neuroscience Center, Department of Molecular and Cellular Neuroscience, La Jolla, CA 92037- high levels of Ab1–40 and Ab1–42 peptides and oligo- 1000, USA mers, recapitulate the reduction of excitatory synapses, exhibit neuronal network dysfunction and cognitive def- Corresponding author: Polleux, Franck ([email protected]) 1 icits in spatial learning [6,11]. -

Developing Biomarkers for Livestock Science
Developing biomarkers for livestock Science Ongoing research and future developments Marinus te Pas Outline . Introduction ● What are biomarkers ● Why do we need them . Examples ● omics levels . The future ● Big data ● Systems biology / Synthetic biology 2 Introduction: What are biomarkers? . Biological processes underlie all livestock (production) traits ● Measure the status of a biological process = know the trait! . Can be any molecule in a cell ● No need to know the causal factor for a trait . Well known example: blood glucose level for diabetes Introduction: Why do we need biomarkers? . The mission of WageningenUR: Sustainably produce enough high quality food for all people on the planet with an ecological footprint as low as possible 4 What can the industry do with biomarkers? . Diagnostic tool ● What is the biological mechanism underlying a trait? . Prediction tool ● What outcome can I expect from an intervention? . Monitoring tool ● What is the actual status of a process? . Speed up your process, improve your traits Some examples * Transcriptomics * Proteomics * Metabolomics Why Biomarkers for meat quality? . Meat quality has low heritability (h2=0.1-0.2) ● Predictive capacity of genetic markers low . High environmental influence ● Feed, animal handling (stress), management (housing), ... Meat quality can only be measured after 1-several days post slaughter . Need to differentiate between retail, processing industry, restaurants, .... Biomarkers can do all that and more faster, predictive, .. Example Transcriptomics biomarkers for meat quality . Pork production chain . Biomarkers for traits . High quality fresh pork . Meat colour N production chain ● A* 14 . German Pietrain ● L* 4 (microarray) ● Reflection 10 . Verification: Danish . Drip loss 2 Yorkshire (PCR) . Ultimate pH 6 . -

Ubiquitylome Profiling of Parkin-Null Brain Reveals Dysregulation Of
Neurobiology of Disease 127 (2019) 114–130 Contents lists available at ScienceDirect Neurobiology of Disease journal homepage: www.elsevier.com/locate/ynbdi Ubiquitylome profiling of Parkin-null brain reveals dysregulation of calcium T homeostasis factors ATP1A2, Hippocalcin and GNA11, reflected by altered firing of noradrenergic neurons Key J.a,1, Mueller A.K.b,1, Gispert S.a, Matschke L.b, Wittig I.c, Corti O.d,e,f,g, Münch C.h, ⁎ ⁎ Decher N.b, , Auburger G.a, a Exp. Neurology, Goethe University Medical School, 60590 Frankfurt am Main, Germany b Institute for Physiology and Pathophysiology, Vegetative Physiology and Marburg Center for Mind, Brain and Behavior - MCMBB; Clinic for Neurology, Philipps-University Marburg, 35037 Marburg, Germany c Functional Proteomics, SFB 815 Core Unit, Goethe University Medical School, 60590 Frankfurt am Main, Germany d Institut du Cerveau et de la Moelle épinière, ICM, Paris, F-75013, France e Inserm, U1127, Paris, F-75013, France f CNRS, UMR 7225, Paris, F-75013, France g Sorbonne Universités, Paris, F-75013, France h Institute of Biochemistry II, Goethe University Medical School, 60590 Frankfurt am Main, Germany ARTICLE INFO ABSTRACT Keywords: Parkinson's disease (PD) is the second most frequent neurodegenerative disorder in the old population. Among Parkinson's disease its monogenic variants, a frequent cause is a mutation in the Parkin gene (Prkn). Deficient function of Parkin Mitochondria triggers ubiquitous mitochondrial dysfunction and inflammation in the brain, but it remains unclear howse- Parkin lective neural circuits become vulnerable and finally undergo atrophy. Ubiquitin We attempted to go beyond previous work, mostly done in peripheral tumor cells, which identified protein Calcium targets of Parkin activity, an ubiquitin E3 ligase. -

Saracatinib Is an Efficacious Clinical Candidate for Fibrodysplasia Ossificans Progressiva
RESEARCH ARTICLE Saracatinib is an efficacious clinical candidate for fibrodysplasia ossificans progressiva Eleanor Williams,1 Jana Bagarova,2 Georgina Kerr,1 Dong-Dong Xia,2 Elsie S. Place,3 Devaveena Dey,2 Yue Shen,2 Geoffrey A. Bocobo,2 Agustin H. Mohedas,2 Xiuli Huang,4 Philip E. Sanderson,4 Arthur Lee,4 Wei Zheng,4 Aris N. Economides,5 James C. Smith,3 Paul B. Yu,2 and Alex N. Bullock1 1Centre for Medicines Discovery, University of Oxford, Oxford, United Kingdom. 2Department of Medicine, Cardiovascular Division, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 3Developmental Biology Laboratory, Francis Crick Institute, London, United Kingdom. 4National Center for Advancing Translational Sciences, NIH, Bethesda, Maryland, USA. 5Regeneron Pharmaceuticals Inc., Tarrytown, New York, USA. Currently, no effective therapies exist for fibrodysplasia ossificans progressiva (FOP), a rare congenital syndrome in which heterotopic bone is formed in soft tissues owing to dysregulated activity of the bone morphogenetic protein (BMP) receptor kinase ALK2 (also known as ACVR1). From a screen of known biologically active compounds, we identified saracatinib as a potent ALK2 kinase inhibitor. In enzymatic and cell-based assays, saracatinib preferentially inhibited ALK2, compared with other receptors of the BMP/TGF-β signaling pathway, and induced dorsalization in zebrafish embryos consistent with BMP antagonism. We further tested the efficacy of saracatinib using an inducible ACVR1Q207D-transgenic mouse line, which provides a model of heterotopic ossification (HO), as well as an inducible ACVR1R206H-knockin mouse, which serves as a genetically and physiologically faithful FOP model. In both models, saracatinib was well tolerated and potently inhibited the development of HO, even when administered transiently following soft tissue injury. -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -
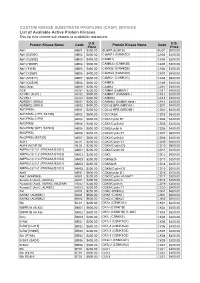
CUSTOM KINASE SUBSTRATE PROFILING (CKSP) SERVICE List of Available Active Protein Kinases This List May Change with Respect to Availability and Pricing
CUSTOM KINASE SUBSTRATE PROFILING (CKSP) SERVICE List of Available Active Protein Kinases This list may change with respect to availability and pricing. Protein Kinase Name Code U.S. Protein Kinase Name Code U.S. Price Price Abl1 AB01 $200.00 BUBR1(BUB1B) BU01 $600.00 Abl1 [E255K] AB02 $400.00 CaMK1δ (CAMK1D) CA03 $200.00 Abl1 [G250E] AB03 $400.00 CAMK1γ CA04 $200.00 Abl1 [H369P] AB04 $600.00 CAMK2α (CAMK2B) CA05 $200.00 Abl1 [T315I] AB05 $400.00 CaMK2β (CAMK2B) CA06 $200.00 Abl1 [Y253F] AB06 $400.00 CaMK2δ (CAMK2D) CA07 $400.00 Abl1 [M351T] AB07 $600.00 CaMK2γ (CAMK2G) CA08 $600.00 Abl1 [Q252H] AB08 $600.00 CAMK3γ CA09 $200.00 Abl2 (Arg) AB09 $200.00 CAMK4 CA10 $200.00 ACK AC01 $200.00 CAMK4 (CaMKIV ) CA11 $400.00 ACVR1 (ALK2 ) AC02 $400.00 CAMKK1 (CAMKKA ) CA12 $200.00 ACVRL1 AC03 $400.00 CAMKK2 CA13 $200.00 ADRBK1 (GRK2) AD01 $200.00 CAMKK2 (CaMKK beta ) CA14 $400.00 ADRBK2 (GRK3) AD02 $400.00 CDC42 BPA (MRCKA ) CD01 $400.00 Akt1/PKBα AK01 $200.00 CDC42 BPB (MRCKB ) CD02 $400.00 Akt1/PKBα [δPH, S473D] AK02 $600.00 CDC7/ASK CD03 $600.00 Akt1/PKBα [δPH] AK03 $600.00 CDK1/cyclin B1 CD04 $400.00 Akt2/PKBβ AK04 $200.00 CDK1/CyclinA2 CD05 $200.00 Akt2/PKBβ [δPH, S474D] AK05 $600.00 CDK2/cyclin A CD06 $400.00 Akt3/PKBγ AK06 $200.00 CDK2/Cyclin E1 CD07 $600.00 Akt3/PKBγ [S472D] AK07 $600.00 CDK2/CyclinA2 CD08 $200.00 ALK1 AL01 $200.00 CDK3/Cyclin E1 CD09 $600.00 ALK4 (ACVR1B) AL02 $200.00 CDK4//Cyclin D3 CD10 $600.00 AMPKα1/β1/γ1 (PRKAA1/B1/G1) AM01 $200.00 CDK4/Cyclin D1 CD11 $200.00 AMPKα1/β1/γ2 (PRKAA1/B1/G2) AM02 $200.00 CDK5 CD12 $600.00 -

Supplementary Table 1
SI Table S1. Broad protein kinase selectivity for PF-2771. Kinase, PF-2771 % Inhibition at 10 μM Service Kinase, PF-2771 % Inhibition at 1 μM Service rat RPS6KA1 (RSK1) 39 Dundee AURKA (AURA) 24 Invitrogen IKBKB (IKKb) 26 Dundee CDK2 /CyclinA 21 Invitrogen mouse LCK 25 Dundee rabbit MAP2K1 (MEK1) 19 Dundee AKT1 (AKT) 21 Dundee IKBKB (IKKb) 16 Dundee CAMK1 (CaMK1a) 19 Dundee PKN2 (PRK2) 14 Dundee RPS6KA5 (MSK1) 18 Dundee MAPKAPK5 14 Dundee PRKD1 (PKD1) 13 Dundee PIM3 12 Dundee MKNK2 (MNK2) 12 Dundee PRKD1 (PKD1) 12 Dundee MARK3 10 Dundee NTRK1 (TRKA) 12 Invitrogen SRPK1 9 Dundee MAPK12 (p38g) 11 Dundee MAPKAPK5 9 Dundee MAPK8 (JNK1a) 11 Dundee MAPK13 (p38d) 8 Dundee rat PRKAA2 (AMPKa2) 11 Dundee AURKB (AURB) 5 Dundee NEK2 11 Invitrogen CSK 5 Dundee CHEK2 (CHK2) 11 Invitrogen EEF2K (EEF-2 kinase) 4 Dundee MAPK9 (JNK2) 9 Dundee PRKCA (PKCa) 4 Dundee rat RPS6KA1 (RSK1) 8 Dundee rat PRKAA2 (AMPKa2) 4 Dundee DYRK2 7 Dundee rat CSNK1D (CKId) 3 Dundee AKT1 (AKT) 7 Dundee LYN 3 BioPrint PIM2 7 Invitrogen CSNK2A1 (CKIIa) 3 Dundee MAPK15 (ERK7) 6 Dundee CAMKK2 (CAMKKB) 1 Dundee mouse LCK 5 Dundee PIM3 1 Dundee PDPK1 (PDK1) (directed 5 Invitrogen rat DYRK1A (MNB) 1 Dundee RPS6KB1 (p70S6K) 5 Dundee PBK 0 Dundee CSNK2A1 (CKIIa) 4 Dundee PIM1 -1 Dundee CAMKK2 (CAMKKB) 4 Dundee DYRK2 -2 Dundee SRC 4 Invitrogen MAPK12 (p38g) -2 Dundee MYLK2 (MLCK_sk) 3 Invitrogen NEK6 -3 Dundee MKNK2 (MNK2) 2 Dundee RPS6KB1 (p70S6K) -3 Dundee SRPK1 2 Dundee AKT2 -3 Dundee MKNK1 (MNK1) 2 Dundee RPS6KA3 (RSK2) -3 Dundee CHEK1 (CHK1) 2 Invitrogen rabbit MAP2K1 (MEK1) -4 Dundee -

ACVR1C Antibody Cat
ACVR1C Antibody Cat. No.: 4795 ACVR1C Antibody Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human, Mouse, Rat ACVR1C antibody was raised against a 15 amino acid synthetic peptide near the amino terminus of the human ACVR1C. IMMUNOGEN: The immunogen is located within amino acids 130 - 180 of ACVR1C. TESTED APPLICATIONS: ELISA, WB ACVR1C antibody can be used for detection of ACVR1C by Western blot at 1 and 2 μg/mL. APPLICATIONS: Antibody validated: Western Blot in human samples. All other applications and species not yet tested. SPECIFICITY: This antibody is predicted to have no cross-reactivity to ACVR1 or ACVR1B. POSITIVE CONTROL: 1) Cat. No. 1309 - Human Placenta Tissue Lysate Properties PURIFICATION: ACVR1C Antibody is affinity chromatography purified via peptide column. CLONALITY: Polyclonal September 25, 2021 1 https://www.prosci-inc.com/acvr1c-antibody-4795.html ISOTYPE: IgG CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: ACVR1C Antibody is supplied in PBS containing 0.02% sodium azide. CONCENTRATION: 1 mg/mL ACVR1C antibody can be stored at 4˚C for three months and -20˚C, stable for up to one STORAGE CONDITIONS: year. As with all antibodies care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: ACVR1 ACVR1C Antibody: FOP, ALK2, SKR1, TSRI, ACTRI, ACVR1A, ACVRLK2, Activin receptor ALTERNATE NAMES: type-1, Activin receptor type I, ACTR-I ACCESSION NO.: Q8NER5 PROTEIN GI NO.: 4501895 GENE ID: 90 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References ACVR1C Antibody: Activins are dimeric growth and differentiation factors which belong to the transforming growth factor-beta (TGF-beta) superfamily of structurally related signaling proteins. -

Human ALK-7 / ACVR1C Protein (ECD, Fc Tag)
Human ALK-7 / ACVR1C Protein (ECD, Fc Tag) Catalog Number: 10869-H02H General Information SDS-PAGE: Gene Name Synonym: ACVRLK7; ALK7 Protein Construction: A DNA sequence encoding the human ACVR1C (NP_660302.2) (Met1- Glu113) was expressed with the Fc region of human IgG1 at the C- terminus. Source: Human Expression Host: HEK293 Cells QC Testing Purity: > 95 % as determined by SDS-PAGE. Endotoxin: Protein Description < 1.0 EU per μg protein as determined by the LAL method. ALK-7, also known as ALK7 and ACVR1C, belongs to the ALK family. It is a type I receptor for the TGFB family of signaling molecules. TGF-β is the Stability: prototype of a protein superfamily which, in humans, contains at least 35 members, including activins, inhibins, bone morphogenetic proteins, Samples are stable for up to twelve months from date of receipt at -70 ℃ growth/differentiation factors, and Müllerian inhibiting substance. ALK-7 is a serine-threonine kinase that can cause the activation of one of the SMAD Predicted N terminal: Leu 22 signal transducers, SMAD2. ALK-7 has a ligand known as Nodal. Nodal Molecular Mass: stimulates the secretion of TIMP-1 and inhibits matrix metalloproteinases MMP-2 and MMP-9 activity. The overexpression of Nodal or constitutively The recombinant human ACVR1C consists 330 amino acids and predicts active ALK-7 decreases cell migration and invasion, whereas knock-down a molecular mass of 36.6 kDa. of Nodal and ALK-7 has the opposite effects. Formulation: References Lyophilized from sterile PBS, pH 7.4. 1.Lin YY, et al. (2012) Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. -

140503 IPF Signatures Supplement Withfigs Thorax
Supplementary material for Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis Daryle J. DePianto1*, Sanjay Chandriani1⌘*, Alexander R. Abbas1, Guiquan Jia1, Elsa N. N’Diaye1, Patrick Caplazi1, Steven E. Kauder1, Sabyasachi Biswas1, Satyajit K. Karnik1#, Connie Ha1, Zora Modrusan1, Michael A. Matthay2, Jasleen Kukreja3, Harold R. Collard2, Jackson G. Egen1, Paul J. Wolters2§, and Joseph R. Arron1§ 1Genentech Research and Early Development, South San Francisco, CA 2Department of Medicine, University of California, San Francisco, CA 3Department of Surgery, University of California, San Francisco, CA ⌘Current address: Novartis Institutes for Biomedical Research, Emeryville, CA. #Current address: Gilead Sciences, Foster City, CA. *DJD and SC contributed equally to this manuscript §PJW and JRA co-directed this project Address correspondence to Paul J. Wolters, MD University of California, San Francisco Department of Medicine Box 0111 San Francisco, CA 94143-0111 [email protected] or Joseph R. Arron, MD, PhD Genentech, Inc. MS 231C 1 DNA Way South San Francisco, CA 94080 [email protected] 1 METHODS Human lung tissue samples Tissues were obtained at UCSF from clinical samples from IPF patients at the time of biopsy or lung transplantation. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to criteria established by the consensus classification of the American Thoracic Society (ATS) and European Respiratory Society (ERS), Japanese Respiratory Society (JRS), and the Latin American Thoracic Association (ALAT) (ref. 5 in main text). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network.