Novel Cell Types and Developmental Lineages Revealed by Single-Cell
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deregulated Gene Expression Pathways in Myelodysplastic Syndrome Hematopoietic Stem Cells
Leukemia (2010) 24, 756–764 & 2010 Macmillan Publishers Limited All rights reserved 0887-6924/10 $32.00 www.nature.com/leu ORIGINAL ARTICLE Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells A Pellagatti1, M Cazzola2, A Giagounidis3, J Perry1, L Malcovati2, MG Della Porta2,MJa¨dersten4, S Killick5, A Verma6, CJ Norbury7, E Hellstro¨m-Lindberg4, JS Wainscoat1 and J Boultwood1 1LRF Molecular Haematology Unit, NDCLS, John Radcliffe Hospital, Oxford, UK; 2Department of Hematology Oncology, University of Pavia Medical School, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy; 3Medizinische Klinik II, St Johannes Hospital, Duisburg, Germany; 4Division of Hematology, Department of Medicine, Karolinska Institutet, Stockholm, Sweden; 5Department of Haematology, Royal Bournemouth Hospital, Bournemouth, UK; 6Albert Einstein College of Medicine, Bronx, NY, USA and 7Sir William Dunn School of Pathology, University of Oxford, Oxford, UK To gain insight into the molecular pathogenesis of the the World Health Organization.6,7 Patients with refractory myelodysplastic syndromes (MDS), we performed global gene anemia (RA) with or without ringed sideroblasts, according to expression profiling and pathway analysis on the hemato- poietic stem cells (HSC) of 183 MDS patients as compared with the the French–American–British classification, were subdivided HSC of 17 healthy controls. The most significantly deregulated based on the presence or absence of multilineage dysplasia. In pathways in MDS include interferon signaling, thrombopoietin addition, patients with RA with excess blasts (RAEB) were signaling and the Wnt pathways. Among the most signifi- subdivided into two categories, RAEB1 and RAEB2, based on the cantly deregulated gene pathways in early MDS are immuno- percentage of bone marrow blasts. -

Neuroinflammation Triggered by Β-Glucan/Dectin-1 Signaling Enables CNS Axon Regeneration
Neuroinflammation triggered by β-glucan/dectin-1 signaling enables CNS axon regeneration Katherine T. Baldwina,b,1, Kevin S. Carbajalc,d,1, Benjamin M. Segalc,d,2, and Roman J. Gigera,b,c,d,2 aDepartment of Cell and Developmental Biology, bCellular and Molecular Biology Graduate Program, cNeuroscience Graduate Program, and dHoltom-Garrett Program in Neuroimmunology, Department of Neurology, University of Michigan School of Medicine, Ann Arbor, MI 48109 Edited by Ben A. Barres, Stanford University School of Medicine, Stanford, CA, and approved January 22, 2015 (received for review December 22, 2014) Innate immunity can facilitate nervous system regeneration, yet growth can be undermined by concurrent toxicity (9). A deeper the underlying cellular and molecular mechanisms are not well understanding of these opposing effects will be important for understood. Here we show that intraocular injection of lipopoly- exploiting immunomodulatory pathways to promote neural re- saccharide (LPS), a bacterial cell wall component, or the fungal cell pair while minimizing bystander damage. wall extract zymosan both lead to rapid and comparable intravi- In the present study, we investigated the pathways that drive treal accumulation of blood-derived myeloid cells. However, when innate immune-mediated axon regeneration after ONC. We in- combined with retro-orbital optic nerve crush injury, lengthy duced sterile inflammation in the vitreous on the day of injury by growth of severed retinal ganglion cell (RGC) axons occurs only i.o. administration of zymosan or constituents of zymosan clas- in zymosan-injected mice, and not in LPS-injected mice. In mice sified as pathogen-associated molecular patterns (PAMPs). PAMPs dectin-1 deficient for the pattern recognition receptor but not are highly conserved microbial structures that serve as ligands for TLR2 , Toll-like receptor-2 ( ) zymosan-mediated RGC regeneration pattern recognition receptors (PRRs). -

Identification of Genes Concordantly Expressed with Atoh1 During Inner Ear Development
Original Article doi: 10.5115/acb.2011.44.1.69 pISSN 2093-3665 eISSN 2093-3673 Identification of genes concordantly expressed with Atoh1 during inner ear development Heejei Yoon, Dong Jin Lee, Myoung Hee Kim, Jinwoong Bok Department of Anatomy, Brain Korea 21 Project for Medical Science, College of Medicine, Yonsei University, Seoul, Korea Abstract: The inner ear is composed of a cochlear duct and five vestibular organs in which mechanosensory hair cells play critical roles in receiving and relaying sound and balance signals to the brain. To identify novel genes associated with hair cell differentiation or function, we analyzed an archived gene expression dataset from embryonic mouse inner ear tissues. Since atonal homolog 1a (Atoh1) is a well known factor required for hair cell differentiation, we searched for genes expressed in a similar pattern with Atoh1 during inner ear development. The list from our analysis includes many genes previously reported to be involved in hair cell differentiation such as Myo6, Tecta, Myo7a, Cdh23, Atp6v1b1, and Gfi1. In addition, we identified many other genes that have not been associated with hair cell differentiation, including Tekt2, Spag6, Smpx, Lmod1, Myh7b, Kif9, Ttyh1, Scn11a and Cnga2. We examined expression patterns of some of the newly identified genes using real-time polymerase chain reaction and in situ hybridization. For example, Smpx and Tekt2, which are regulators for cytoskeletal dynamics, were shown specifically expressed in the hair cells, suggesting a possible role in hair cell differentiation or function. Here, by re- analyzing archived genetic profiling data, we identified a list of novel genes possibly involved in hair cell differentiation. -

Porichthys Notatus), a Teleost with Divergent Sexual Phenotypes
Portland State University PDXScholar Biology Faculty Publications and Presentations Biology 11-11-2015 Saccular Transcriptome Profiles of the Seasonal Breeding Plainfin Midshipman Fish (Porichthys notatus), a Teleost with Divergent Sexual Phenotypes Joshua J. Faber-Hammond Portland State University Manoj P. Samanta Systemix Institute Elizabeth A. Whitchurch Humboldt State University Dustin Manning Oregon Health & Science University Joseph A. Sisneros University of Washington Follow this and additional works at: https://pdxscholar.library.pdx.edu/bio_fac Part of the Biology Commons LetSee next us page know for additional how authors access to this document benefits ou.y Citation Details Faber-Hammond, J., Samanta, M. P., Whitchurch, E. A., Manning, D., Sisneros, J. A., & Coffin, A. B. (2015). Saccular Transcriptome Profiles of the Seasonal Breeding Plainfin Midshipman Fish (Porichthys notatus), a Teleost with Divergent Sexual Phenotypes. PloS One, 10(11), e0142814. This Article is brought to you for free and open access. It has been accepted for inclusion in Biology Faculty Publications and Presentations by an authorized administrator of PDXScholar. Please contact us if we can make this document more accessible: [email protected]. Authors Joshua J. Faber-Hammond, Manoj P. Samanta, Elizabeth A. Whitchurch, Dustin Manning, Joseph A. Sisneros, and Allison B. Coffin This article is available at PDXScholar: https://pdxscholar.library.pdx.edu/bio_fac/108 RESEARCH ARTICLE Saccular Transcriptome Profiles of the Seasonal Breeding Plainfin Midshipman -

Figure S1. Representative Report Generated by the Ion Torrent System Server for Each of the KCC71 Panel Analysis and Pcafusion Analysis
Figure S1. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. A Figure S1. Continued. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. B Figure S2. Comparative analysis of the variant frequency found by the KCC71 panel and calculated from publicly available cBioPortal datasets. For each of the 71 genes in the KCC71 panel, the frequency of variants was calculated as the variant number found in the examined cases. Datasets marked with different colors and sample numbers of prostate cancer are presented in the upper right. *Significantly high in the present study. Figure S3. Seven subnetworks extracted from each of seven public prostate cancer gene networks in TCNG (Table SVI). Blue dots represent genes that include initial seed genes (parent nodes), and parent‑child and child‑grandchild genes in the network. Graphical representation of node‑to‑node associations and subnetwork structures that differed among and were unique to each of the seven subnetworks. TCNG, The Cancer Network Galaxy. Figure S4. REVIGO tree map showing the predicted biological processes of prostate cancer in the Japanese. Each rectangle represents a biological function in terms of a Gene Ontology (GO) term, with the size adjusted to represent the P‑value of the GO term in the underlying GO term database. -

Saccule and Utricle
THE SPECIAL SENSES VESTIBULAR FUNCTION DR SYED SHAHID HABIB MBBS DSDM PGDCR FCPS Professor Dept. of Physiology College of Medicine & KKUH OBJECTIVES At the end of this lecture you should be able to describe: Functional anatomy of Vestibular apparatus Dynamic and static equilibrium Role of utricle and saccule in linear acceleration Role of semicircular canals in angular motions Vestibular Reflexes Overview of Static Proprioception & Balance position sense (Ia) Dynamic position sense (II) Static Equilibrium Utricle & Saccule Neck Proprioceptors Linear Acceleration Horizontal (Utricle) Visual Information (vesitbulo Ocular) Linear Acceleration Vestibular Apparatus Horizontal (Saccule) Proprioception Chest Wall Equilibrium Angular Acceleration Proprioceptors (SCCs) air pressure against body Predictive Functions (SCCs) Footpads pressure To balance the centre of gravity must be above the support point. Centre of gravity Physiology Of Body Balance Balance & Equilibrium Balance is the ability to maintain the equilibrium of the body • Foot position affects standing balance Equilibrium is the state of a body or physical system at rest or in un accelerated motion in which the resultant of all forces acting on it is zero and the sum of all torques about any axis is zero. There are 2 types of Equilibrium » Static - » Dynamic – Static Equilibrium keep the body in a desired position Static equilibrium –The equilibrium is maintained in a FIXED POSITION, usually while stood on one foot or maintenance of body posture relative to gravity while the body is still. Dynamic Equilibrium to move the body in a controlled way Dynamic equilibrium The equilibrium must be maintained while performing a task which involves MOVEMENT e.g. Walking the beam. -

Preclinical Evaluation of Protein Disulfide Isomerase Inhibitors for the Treatment of Glioblastoma by Andrea Shergalis
Preclinical Evaluation of Protein Disulfide Isomerase Inhibitors for the Treatment of Glioblastoma By Andrea Shergalis A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Medicinal Chemistry) in the University of Michigan 2020 Doctoral Committee: Professor Nouri Neamati, Chair Professor George A. Garcia Professor Peter J. H. Scott Professor Shaomeng Wang Andrea G. Shergalis [email protected] ORCID 0000-0002-1155-1583 © Andrea Shergalis 2020 All Rights Reserved ACKNOWLEDGEMENTS So many people have been involved in bringing this project to life and making this dissertation possible. First, I want to thank my advisor, Prof. Nouri Neamati, for his guidance, encouragement, and patience. Prof. Neamati instilled an enthusiasm in me for science and drug discovery, while allowing me the space to independently explore complex biochemical problems, and I am grateful for his kind and patient mentorship. I also thank my committee members, Profs. George Garcia, Peter Scott, and Shaomeng Wang, for their patience, guidance, and support throughout my graduate career. I am thankful to them for taking time to meet with me and have thoughtful conversations about medicinal chemistry and science in general. From the Neamati lab, I would like to thank so many. First and foremost, I have to thank Shuzo Tamara for being an incredible, kind, and patient teacher and mentor. Shuzo is one of the hardest workers I know. In addition to a strong work ethic, he taught me pretty much everything I know and laid the foundation for the article published as Chapter 3 of this dissertation. The work published in this dissertation really began with the initial identification of PDI as a target by Shili Xu, and I am grateful for his advice and guidance (from afar!). -
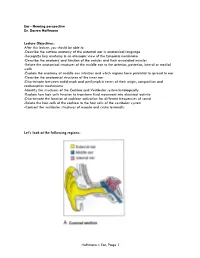
Ear, Page 1 Lecture Outline
Ear - Hearing perspective Dr. Darren Hoffmann Lecture Objectives: After this lecture, you should be able to: -Describe the surface anatomy of the external ear in anatomical language -Recognize key anatomy in an otoscopic view of the tympanic membrane -Describe the anatomy and function of the ossicles and their associated muscles -Relate the anatomical structures of the middle ear to the anterior, posterior, lateral or medial walls -Explain the anatomy of middle ear infection and which regions have potential to spread to ear -Describe the anatomical structures of the inner ear -Discriminate between endolymph and perilymph in terms of their origin, composition and reabsorption mechanisms -Identify the structures of the Cochlea and Vestibular system histologically -Explain how hair cells function to transform fluid movement into electrical activity -Discriminate the location of cochlear activation for different frequencies of sound -Relate the hair cells of the cochlea to the hair cells of the vestibular system -Contrast the vestibular structures of macula and crista terminalis Let’s look at the following regions: Hoffmann – Ear, Page 1 Lecture Outline: C1. External Ear Function: Amplification of Sound waves Parts Auricle Visible part of external ear (pinna) Helix – large outer rim Tragus – tab anterior to external auditory meatus External auditory meatus Auditory Canal/External Auditory Meatus Leads from Auricle to Tympanic membrane Starts cartilaginous, becomes bony as it enters petrous part of temporal bone Earwax (Cerumen) Complex mixture -

Loss of the Tectorial Membrane Protein CEACAM16 Enhances Spontaneous, Stimulus-Frequency, and Transiently Evoked Otoacoustic Emissions
The Journal of Neuroscience, July 30, 2014 • 34(31):10325–10338 • 10325 Cellular/Molecular Loss of the Tectorial Membrane Protein CEACAM16 Enhances Spontaneous, Stimulus-Frequency, and Transiently Evoked Otoacoustic Emissions Mary Ann Cheatham,1 Richard J. Goodyear,3 Kazuaki Homma,4 P. Kevin Legan,3 Julia Korchagina,3 Souvik Naskar,3 Jonathan H. Siegel,1 X Peter Dallos,1,2 Jing Zheng,4 and Guy P. Richardson3 1Roxelyn and Richard Pepper Department of Communication Sciences and Disorders, The Knowles Hearing Center, and 2Department of Neurobiology, Northwestern University, Evanston, Illinois 60208, 3Sussex Neuroscience, School of Life Sciences, University of Sussex, Falmer, Brighton, BN1 9QG, United Kingdom, and 4Department of Otolaryngology-Head and Neck Surgery, The Knowles Hearing Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois 60611 ␣-Tectorin (TECTA), -tectorin (TECTB), and carcinoembryonic antigen-related cell adhesion molecule 16 (CEACAM) are secreted glycoproteins that are present in the tectorial membrane (TM), an extracellular structure overlying the hearing organ of the inner ear, the organ of Corti. Previous studies have shown that TECTA and TECTB are both required for formation of the striated-sheet matrix within which collagen fibrils of the TM are imbedded and that CEACAM16 interacts with TECTA. To learn more about the structural and functional significance of CEACAM16, we created a Ceacam16-null mutant mouse. In the absence of CEACAM16, TECTB levels are reduced, a clearly defined striated-sheet matrix does not develop, and Hensen’s stripe, a prominent feature in the basal two-thirds of the TM in WT mice, is absent. CEACAM16 is also shown to interact with TECTB, indicating that it may stabilize interactions between TECTA and TECTB. -

Role of Amylase in Ovarian Cancer Mai Mohamed University of South Florida, [email protected]
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School July 2017 Role of Amylase in Ovarian Cancer Mai Mohamed University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the Pathology Commons Scholar Commons Citation Mohamed, Mai, "Role of Amylase in Ovarian Cancer" (2017). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/6907 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Role of Amylase in Ovarian Cancer by Mai Mohamed A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Pathology and Cell Biology Morsani College of Medicine University of South Florida Major Professor: Patricia Kruk, Ph.D. Paula C. Bickford, Ph.D. Meera Nanjundan, Ph.D. Marzenna Wiranowska, Ph.D. Lauri Wright, Ph.D. Date of Approval: June 29, 2017 Keywords: ovarian cancer, amylase, computational analyses, glycocalyx, cellular invasion Copyright © 2017, Mai Mohamed Dedication This dissertation is dedicated to my parents, Ahmed and Fatma, who have always stressed the importance of education, and, throughout my education, have been my strongest source of encouragement and support. They always believed in me and I am eternally grateful to them. I would also like to thank my brothers, Mohamed and Hussien, and my sister, Mariam. I would also like to thank my husband, Ahmed. -

Mathematical Model of the Cupula-Endolymph System with Morphological Parameters for the Axolotl (Ambystoma Tigrinum) Semicircular Canals
138 The Open Medical Informatics Journal, 2008, 2, 138-148 Open Access Mathematical Model of the Cupula-Endolymph System with Morphological Parameters for the Axolotl (Ambystoma tigrinum) Semicircular Canals Rosario Vega1, Vladimir V. Alexandrov2,3, Tamara B. Alexandrova1,3 and Enrique Soto*,1 1Instituto de Fisiología, Universidad Autónoma de Puebla, 2Facultad de Ciencias Físico Matemáticas, Universidad Autónoma de Puebla, 3 Lomonosov Moscow State University, Mexico Abstract: By combining mathematical methods with the morphological analysis of the semicircular canals of the axolotl (Ambystoma tigrinum), a system of differential equations describing the mechanical coupling in the semicircular canals was obtained. The coefficients of this system have an explicit physiological meaning that allows for the introduction of morphological and dynamical parameters directly into the differential equations. The cupula of the semicircular canals was modeled both as a piston and as a membrane (diaphragm like), and the duct canals as toroids with two main regions: i) the semicircular canal duct and, ii) a larger diameter region corresponding to the ampulla and the utricle. The endolymph motion was described by the Navier-Stokes equations. The analysis of the model demonstrated that cupular behavior dynamics under periodic stimulation is equivalent in both the piston and the membrane cupular models, thus a general model in which the detailed cupular structure is not relevant was derived. Keywords: Inner ear, vestibular, hair cell, transduction, sensory coding, physiology. 1. INTRODUCTION linear acceleration detectors, and the SCs as angular accel- eration detectors, notwithstanding that both sensory organs The processing of sensory information in the semicircular are based on a very similar sensory cell type. -

Molecular Associations and Clinical Significance of Raps In
ORIGINAL RESEARCH published: 21 June 2021 doi: 10.3389/fmolb.2021.677979 Molecular Associations and Clinical Significance of RAPs in Hepatocellular Carcinoma Sarita Kumari 1†, Mohit Arora 2†, Jay Singh 1, Lokesh K. Kadian 2, Rajni Yadav 3, Shyam S. Chauhan 2* and Anita Chopra 1* 1Laboratory Oncology Unit, Dr. BRA-IRCH, All India Institute of Medical Sciences, New Delhi, India, 2Department of Biochemistry, All India Institute of Medical Sciences, New Delhi, India, 3Department of Pathology, All India Institute of Medical Sciences, New Delhi, India Hepatocellular carcinoma (HCC) is an aggressive gastrointestinal malignancy with a high rate of mortality. Multiple studies have individually recognized members of RAP gene family as critical regulators of tumor progression in several cancers, including hepatocellular carcinoma. These studies suffer numerous limitations including a small sample size and lack of analysis of various clinicopathological and molecular Edited by: Veronica Aran, features. In the current study, we utilized authoritative multi-omics databases to Instituto Estadual do Cérebro Paulo determine the association of RAP gene family expression and detailed molecular and Niemeyer (IECPN), Brazil clinicopathological features in hepatocellular carcinoma (HCC). All five RAP genes Reviewed by: were observed to harbor dysregulated expression in HCC compared to normal liver Pooja Panwalkar, Weill Cornell Medicine, United States tissues. RAP2A exhibited strongest ability to differentiate tumors from the normal Jasminka Omerovic, tissues. RAP2A expression was associated with progressive tumor grade, TP53 and University of Split, Croatia CTNNB1 mutation status. Additionally, RAP2A expression was associated with the *Correspondence: Anita Chopra alteration of its copy numbers and DNA methylation. RAP2A also emerged as an [email protected] independent marker for patient prognosis.