A Mutation in the Putative MLH3 Endonuclease Domain Confers a Defect in Both Mismatch Repair and Meiosis in Saccharomyces Cerevisiae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Helicase Mechanisms During Homologous Recombination in Saccharomyces Cerevisiae
BB48CH11_Greene ARjats.cls April 18, 2019 12:24 Annual Review of Biophysics Helicase Mechanisms During Homologous Recombination in Saccharomyces cerevisiae J. Brooks Crickard and Eric C. Greene Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA; email: [email protected], [email protected] Annu. Rev. Biophys. 2019. 48:255–73 Keywords First published as a Review in Advance on homologous recombination, helicase, Srs2, Sgs1, Rad54 March 11, 2019 Access provided by 68.175.70.229 on 06/02/20. For personal use only. The Annual Review of Biophysics is online at Abstract Annu. Rev. Biophys. 2019.48:255-273. Downloaded from www.annualreviews.org biophys.annualreviews.org Helicases are enzymes that move, manage, and manipulate nucleic acids. https://doi.org/10.1146/annurev-biophys-052118- They can be subdivided into six super families and are required for all aspects 115418 of nucleic acid metabolism. In general, all helicases function by converting Copyright © 2019 by Annual Reviews. the chemical energy stored in the bond between the gamma and beta phos- All rights reserved phates of adenosine triphosphate into mechanical work, which results in the unidirectional movement of the helicase protein along one strand of a nu- cleic acid. The results of this translocation activity can range from separation of strands within duplex nucleic acids to the physical remodeling or removal of nucleoprotein complexes. In this review, we focus on describing key heli- cases from the model organism Saccharomyces cerevisiae that contribute to the regulation of homologous recombination, which is an essential DNA repair pathway for fxing damaged chromosomes. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

A Genome-Wide Screen for Genes Affecting Spontaneous Direct-Repeat Recombination In
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.11.943795; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A genome-wide screen for genes affecting spontaneous direct-repeat recombination in 2 Saccharomyces cerevisiae 3 4 5 Daniele Novarina*, Ridhdhi Desai†, Jessica A. Vaisica†, Jiongwen Ou†, Mohammed Bellaoui†,1, 6 Grant W. Brown†,2 and Michael Chang*,3 7 8 *European Research Institute for the Biology of Ageing, University of Groningen, University 9 Medical Center Groningen, 9713 AV Groningen, the Netherlands 10 †Department of Biochemistry and Donnelly Centre, University of Toronto, Toronto, ON M5S 11 3E1, Canada 12 13 1Current address: Genetics Unit, Faculty of Medicine and Pharmacy, University Mohammed 14 Premier, Oujda, Morocco 15 16 2Co-corresponding author: Department of Biochemistry and Donnelly Centre, University of 17 Toronto, 160 College Street, Toronto, ON M5S 3E1 Canada. E-mail: [email protected] 18 3Co-corresponding author: European Research Institute for the Biology of Ageing, University of 19 Groningen, University Medical Center Groningen, Antonius Deusinglaan 1, 9713 AV Groningen, 20 the Netherlands. E-mail: [email protected] 21 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.11.943795; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Research in Biology Using Computation Doi
Journal of Bioinformatics and Genomics 2 (7) 2018 RESEARCH IN BIOLOGY USING COMPUTATION DOI: https://doi.org/10.18454/jbg.2018.2.7.1 Grishaeva T.M.* Department of Cytogenetics, Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow, Russian Federation * Correspodning author (grishaeva[at]vigg.ru) Received: 18.03.2018; Accepted: 05.04.2018; Published: 22.05.2018 COMPARATIVE CONSERVATION OF MEIOTIC PROTEINS IN DIFFERENT PHYLOGENETIC LINES OF EUKARYOTES Research article Abstract Motivation: Meiosis — a two-stage process of sex cell division — is served by several hundreds of proteins. A part of them went to eukaryotes from prokaryotes, others appeared in first eukaryotes, and some proteins appeared de novo in multicellular eukaryotes. We compared the conservation of proteins involved in various processes occurring in meiosis. Results: The conservations of five meiotic enzymes (MLH1, MRE11, MSH4, BRCA1, BRCA2) and three silencing markers (histone H2AX, SUMO1, ATR) were compared using a set of bioinformatics methods. Orthologs of these proteins from the proteomes of model species were compared, representing different lines of development of eukaryotes. Among the enzymes, the most conserved is MLH1, which provide correction of mismatch bases, and the least conserved are BRCA1 and BRCA2 repair enzymes which are present only in vertebrates. Among silencing proteins, histone H2AX is the most conserved one, playing the central part in the regulation of the transcription, in the repair and replication of DNA. The small protein SUMO1, which is involved in many cellular processes, is less conserved. ATR kinase in different species is similar only in the C-terminal part of the molecule. -

Fanconi Anemia, Bloom Syndrome and Breast Cancer
A multiprotein complex in DNA damage response network of Fanconi anemia, Bloom syndrome and Breast cancer Weidong Wang Lab of Genetics, NIA A Multi-protein Complex Connects Two Genomic Instability Diseases: Bloom Syndrome and Fanconi Anemia Bloom Syndrome . Genomic Instability: -sister-chromatid exchange . Cancer predisposition . Mutation in BLM, a RecQ DNA Helicase . BLM participates in: HR-dependent DSB repair Recovery of stalled replication forks . BLM works with Topo IIIa and RMI to Suppress crossover recombination Courtesy of Dr. Ian Hickson A Multi-protein Complex Connects Two Genomic Instability Diseases: Bloom Syndrome and Fanconi Anemia P I l o r t n o BLM IP kDa C HeLa BLAP 250 Nuclear Extract 200- BLM* FANCA* 116- TOPO IIIα* 97- BLAP 100 MLH1* BLM IP BLAP 75 * 66- RPA 70 IgG H 45- * 30- RPA32 IgG L 20- * 12- RPA14 Meetei et al. MCB 2003 A Multi-protein Complex Connects Two Genomic Instability Diseases: Bloom Syndrome and Fanconi Anemia P I A C N A F BLM IP HeLa FANCM= FAAP 250 BLAP 250 Nuclear Extract BLM* BLM* * FANCA* FANCA TOPO IIIα* TOPO IIIα* FAAP 100 BLAP 100 FANCB= FAAP 95 MLH1 FANCA IP BLM IP BLAP 75 BLAP 75 RPA70*/FANCG* RPA 70* FANCC*/FANCE* IgG H FANCL= FAAP 43 FANCF* RPA32* IgG L Meetei et al. MCB 2003 Meetei et al. Nat Genet. 2003, 2004, 2005 BRAFT-a Multisubunit Machine that Maintains Genome Stability and is defective in Fanconi anemia and Bloom syndrome BRAFT Super-complex Fanconi Anemia Bloom Syndrome Core Complex Complex 12 polypeptides 7 polypeptides FANCA BLM Helicase (HJ, fork, D-loop), fork FANCC regression, dHJ dissolution Topo IIIα Topoisomerase, FANCE dHJ dissolution FANCF BLAP75 RMI1 FANCG Stimulates dHJ dissolution. -

Human Muts Homologue MSH4 Physically Interacts with Von Hippel-Lindau Tumor Suppressor-Binding Protein 11
[CANCER RESEARCH 63, 865–872, February 15, 2003] Human MutS Homologue MSH4 Physically Interacts with von Hippel-Lindau Tumor Suppressor-binding Protein 11 Chengtao Her,2 Xiling Wu, Michael D. Griswold, and Feng Zhou School of Molecular Biosciences and Center for Reproductive Biology, Washington State University, Pullman, Washington 99164-4660 [C. H., X. W., M. D. G.], and Bioscience Division, Los Alamos National Laboratory, Los Alamos, New Mexico 87545 [F. Z.] ABSTRACT human MMR genes are linked to the pathogenesis of hereditary nonpolyposis colorectal cancer (HNPCC) and sporadic tumors Increasing evidence indicated that the protein factors involved in DNA associated with microsatellite instability (1). Eukaryotic MutS mismatch repair (MMR) possess meiotic functions beyond the scope of DNA mismatch correction. The important roles of MMR components in homologous proteins MSH2, MSH3, and MSH6 are proteins meiotic processes have been highlighted by the recent identification of two known to participate in DNA MMR through the actions of their additional members of the mammalian MutS homologs, MSH4 and heterodimeric complexes consisting of either MSH2-MSH3 or MSH5. Mammalian MSH4 and MSH5 proteins form a heterodimeric MSH2-MSH6, in which the MSH2-MSH6 heterodimer recognizes complex and play an important role in the meiotic processes. As a step both single-base mismatches and small loops formed by insertions forward to the understanding of the molecular mechanisms underlying or deletions in the DNA, whereas the MSH2-MSH3 heterodimer the roles of these two mammalian MutS homologues, here we have iden- only recognizes small insertions and deletions (3, 4). tified von Hippel-Lindau (VHL) tumor suppressor-binding protein 1 Recent evidence demonstrates that eukaryotes contained a sep- (VBP1) as an interacting protein partner for human MSH4 (hMSH4). -

Genetics of Azoospermia
International Journal of Molecular Sciences Review Genetics of Azoospermia Francesca Cioppi , Viktoria Rosta and Csilla Krausz * Department of Biochemical, Experimental and Clinical Sciences “Mario Serio”, University of Florence, 50139 Florence, Italy; francesca.cioppi@unifi.it (F.C.); viktoria.rosta@unifi.it (V.R.) * Correspondence: csilla.krausz@unifi.it Abstract: Azoospermia affects 1% of men, and it can be due to: (i) hypothalamic-pituitary dysfunction, (ii) primary quantitative spermatogenic disturbances, (iii) urogenital duct obstruction. Known genetic factors contribute to all these categories, and genetic testing is part of the routine diagnostic workup of azoospermic men. The diagnostic yield of genetic tests in azoospermia is different in the different etiological categories, with the highest in Congenital Bilateral Absence of Vas Deferens (90%) and the lowest in Non-Obstructive Azoospermia (NOA) due to primary testicular failure (~30%). Whole- Exome Sequencing allowed the discovery of an increasing number of monogenic defects of NOA with a current list of 38 candidate genes. These genes are of potential clinical relevance for future gene panel-based screening. We classified these genes according to the associated-testicular histology underlying the NOA phenotype. The validation and the discovery of novel NOA genes will radically improve patient management. Interestingly, approximately 37% of candidate genes are shared in human male and female gonadal failure, implying that genetic counselling should be extended also to female family members of NOA patients. Keywords: azoospermia; infertility; genetics; exome; NGS; NOA; Klinefelter syndrome; Y chromosome microdeletions; CBAVD; congenital hypogonadotropic hypogonadism Citation: Cioppi, F.; Rosta, V.; Krausz, C. Genetics of Azoospermia. 1. Introduction Int. J. Mol. Sci. -

Linking the Multiple Functions of Xpf-Ercc1 Endonuclease in Dna Repair to Health Outcomes: Cancer and Aging
LINKING THE MULTIPLE FUNCTIONS OF XPF-ERCC1 ENDONUCLEASE IN DNA REPAIR TO HEALTH OUTCOMES: CANCER AND AGING by Advaitha Madireddy B. Tech, SRM University, India, 2008 Submitted to the Graduate Faculty of Graduate School of Public Health in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2012 UNIVERSITY OF PITTSBURGH Graduate School of Public Health This dissertation was presented by Advaitha Madireddy It was defended on June 25th, 2012 and approved by Candace M. Kammerer, PhD, Assistant Professor, Human Genetics, Graduate School of Public Health, University of Pittsburgh Susanne M. Gollin, PhD, Professor, Human Genetics, Graduate School of Public Health, University of Pittsburgh Patricia L. Opresko, PhD, Assistant Professor, Environmental and Occupational Health, Graduate School of Public Health, University of Pittsburgh Dissertation Advisor: Laura J. Niedernhofer, MD, PhD, Associate Professor, Microbiology and Molecular Genetics, School of Medicine, University of Pitttsburgh ii Copyright © by Advaitha Madireddy 2012 iii LINKING THE MULTIPLE FUNCTIONS OF XPF-ERCC1 ENDONUCLEASE IN DNA REPAIR TO HEALTH OUTCOMES: CANCER AND AGING Advaitha Madireddy, PhD University of Pittsburgh, 2012 XPF-ERCC1 is a structure specific endonuclease in which the XPF subunit is involved in nucleolytic activity and the ERCC1 subunit is involved in DNA binding. They are essential for multiple genome maintenance mechanisms which include the repair of bulky DNA monoadducts via nucleotide excision repair (NER) and also the repair of DNA interstrand crosslinks. In humans, the deficiency of XPF-ERCC1 results in two major syndromes: Xeroderma pigmentosum (XP), characterized by predisposition to skin cancer and XFE, characterized by symptoms of premature aging. -

Suppression of Spontaneous Genome Rearrangements in Yeast DNA Helicase Mutants
Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants Kristina H. Schmidt*†‡ and Richard D. Kolodner*§¶ *Ludwig Institute for Cancer Research and §Departments of Medicine and Cellular and Molecular Medicine and Cancer Center, University of California at San Diego, La Jolla, CA 92093; and †Division of Cell Biology, Microbiology, and Molecular Biology, Department of Biology, University of South Florida, Tampa, FL 33620 Contributed by Richard D. Kolodner, October 2, 2006 (sent for review June 16, 2006) Saccharomyces cerevisiae mutants lacking two of the three DNA the hyperrecombination and DNA-damage sensitive phenotypes of helicases Sgs1, Srs2, and Rrm3 exhibit slow growth that is sup- srs2 mutants are suppressed by HR defects (31). The physical pressed by disrupting homologous recombination. Cells lacking interaction between Srs2 and Pol32, a structural subunit of DNA Sgs1 and Rrm3 accumulate gross-chromosomal rearrangements polymerase delta, suggests that Srs2 may act during DNA replica- (GCRs) that are suppressed by the DNA damage checkpoint and by tion (32). Srs2 also is required for proper activation of Rad53 in homologous recombination-defective mutations. In contrast, rrm3, response to DNA-damaging agents (7, 33), and Srs2 itself is srs2, and srs2 rrm3 mutants have wild-type GCR rates. GCR types in phosphorylated after cells are exposed to methyl-methanesulfon- helicase double mutants include telomere additions, transloca- ate, hydroxyurea, or UV light; however, the significance of this tions, and broken DNAs healed by a complex process of hairpin- phosphorylation is unknown (33). mediated inversion. Spontaneous activation of the Rad53 check- Unlike Sgs1 and Srs2, the Rrm3 helicase has 5Ј-to-3Ј polarity and point kinase in the rrm3 mutant depends on the Mec3͞Rad24 DNA shares homology throughout its helicase domain with the S. -

E2013080118.Full.Pdf
Replication-independent instability of Friedreich’s ataxia GAA repeats during chronological aging Alexander J. Neila,1, Julia A. Hiseya,1, Ishtiaque Quasema, Ryan J. McGintya, Marcin Hitczenkob, Alexandra N. Khristicha, and Sergei M. Mirkina,2 aDepartment of Biology, Tufts University, Medford, MA 02155; and bFederal Reserve Bank of Atlanta, Atlanta, GA 30309 Edited by Sue Jinks-Robertson, Duke University School of Medicine, Durham, NC, and approved December 28, 2020 (received for review June 25, 2020) Nearly 50 hereditary diseases result from the inheritance of abnor- For example, in the case of Huntington’s disease (HD), an in-frame mally long repetitive DNA microsatellites. While it was originally (CAG)n repeat causes the buildup of disruptive polyglutamine believed that the size of inherited repeats is the key factor in protein aggregates in the brain with age (7, 8). In FRDA, where the disease development, it has become clear that somatic instability of causative repetitive element is noncoding, it has been hypothesized these repeats throughout an individual’s lifetime strongly contrib- that frataxin deficiency drives disease progression in a similar utes to disease onset and progression. Importantly, somatic insta- fashion via accumulation of toxic iron due to the dysregulation of bility is commonly observed in terminally differentiated, postmitotic iron homeostasis (9). An emerging alternative to the toxicity hy- cells, such as neurons. To unravel the mechanisms of repeat insta- pothesis is that the primary driver of symptom onset and progres- bility in nondividing cells, we created an experimental system to sion could be age-related, somatic expansions of the inherited ’ analyze the mutability of Friedreich s ataxia (GAA)n repeats during disease-size microsatellite. -

Crossing and Zipping: Molecular Duties of the ZMM Proteins in Meiosis Alexandra Pyatnitskaya, Valérie Borde, Arnaud De Muyt
Crossing and zipping: molecular duties of the ZMM proteins in meiosis Alexandra Pyatnitskaya, Valérie Borde, Arnaud de Muyt To cite this version: Alexandra Pyatnitskaya, Valérie Borde, Arnaud de Muyt. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma, Springer Verlag, 2019, 10.1007/s00412-019-00714-8. hal-02413016 HAL Id: hal-02413016 https://hal.archives-ouvertes.fr/hal-02413016 Submitted on 16 Dec 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Manuscript Click here to access/download;Manuscript;review ZMM Chromosoma2019_Revised#2.docx Click here to view linked References Crossing and zipping: molecular duties of the ZMM proteins in meiosis 1 2 3 1,2 1,2,* 1,2,* 4 Alexandra Pyatnitskaya , Valérie Borde and Arnaud De Muyt 5 1 Institut Curie, PSL Research University, CNRS, UMR3244, Paris, France. 6 7 2 Paris Sorbonne Université, Paris, France. 8 9 *Valérie Borde, [email protected]; Arnaud De Muyt, [email protected] 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Keywords : meiosis, crossover, recombination, synaptonemal complex, ZMM 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 1 65 Abstract 1 2 Accurate segregation of homologous chromosomes during meiosis depends on the ability 3 4 of meiotic cells to promote reciprocal exchanges between parental DNA strands, known 5 as crossovers (COs). -
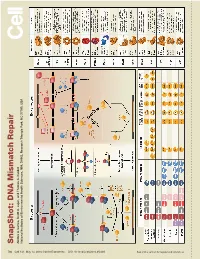
S Na P S H O T: D N a Mism a Tc H R E P a Ir
SnapShot: Repair DNA Mismatch Scott A. Lujan, and Thomas Kunkel A. Larrea, Andres Park, NC 27709, USA Triangle Health Sciences, NIH, DHHS, Research National Institutes of Environmental 730 Cell 141, May 14, 2010 ©2010 Elsevier Inc. DOI 10.1016/j.cell.2010.05.002 See online version for legend and references. SnapShot: DNA Mismatch Repair Andres A. Larrea, Scott A. Lujan, and Thomas A. Kunkel National Institutes of Environmental Health Sciences, NIH, DHHS, Research Triangle Park, NC 27709, USA Mismatch Repair in Bacteria and Eukaryotes Mismatch repair in the bacterium Escherichia coli is initiated when a homodimer of MutS binds as an asymmetric clamp to DNA containing a variety of base-base and insertion-deletion mismatches. The MutL homodimer then couples MutS recognition to the signal that distinguishes between the template and nascent DNA strands. In E. coli, the lack of adenine methylation, catalyzed by the DNA adenine methyltransferase (Dam) in newly synthesized GATC sequences, allows E. coli MutH to cleave the nascent strand. The resulting nick is used for mismatch removal involving the UvrD helicase, single-strand DNA-binding protein (SSB), and excision by single-stranded DNA exonucleases from either direction, depending upon the polarity of the nick relative to the mismatch. DNA polymerase III correctly resynthesizes DNA and ligase completes repair. In bacteria lacking Dam/MutH, as in eukaryotes, the signal for strand discrimination is uncertain but may be the DNA ends associated with replication forks. In these bacteria, MutL harbors a nick-dependent endonuclease that creates a nick that can be used for mismatch excision. Eukaryotic mismatch repair is similar, although it involves several dif- ferent MutS and MutL homologs: MutSα (MSH2/MSH6) recognizes single base-base mismatches and 1–2 base insertion/deletions; MutSβ (MSH2/MSH3) recognizes insertion/ deletion mismatches containing two or more extra bases.