Genetics of Azoospermia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prenatal Diagnosis of Sex Chromosome Mosaicism with Two Marker Chromosomes in Three Cell Lines and a Review of the Literature
MOLECULAR MEDICINE REPORTS 19: 1791-1796, 2019 Prenatal diagnosis of sex chromosome mosaicism with two marker chromosomes in three cell lines and a review of the literature JIANLI ZHENG1, XIAOYU YANG2, HAIYAN LU1, YONGJUAN GUAN1, FANGFANG YANG1, MENGJUN XU1, MIN LI1, XIUQING JI3, YAN WANG3, PING HU3 and YUN ZHOU1 1Department of Prenatal Diagnosis, Laboratory of Clinical Genetics, Maternity and Child Health Care Hospital, Yancheng, Jiangsu 224001; 2Department of Clinical Reproductive Medicine, State Key Laboratory of Reproductive Medicine, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu 210029; 3Department of Prenatal Diagnosis, State Key Laboratory of Reproductive Medicine, Obstetrics and Gynecology Hospital Affiliated to Nanjing Medical University, Nanjing, Jiangsu 210004, P.R. China Received March 31, 2018; Accepted November 21, 2018 DOI: 10.3892/mmr.2018.9798 Abstract. The present study described the diagnosis of a fetus identifying the karyotype, identifying the origin of the marker with sex chromosome mosaicism in three cell lines and two chromosome and preparing effective genetic counseling. marker chromosomes. A 24-year-old woman underwent amniocentesis at 21 weeks and 4 days of gestation due to Introduction noninvasive prenatal testing identifying that the fetus had sex chromosome abnormalities. Amniotic cell culture revealed a Abnormalities involving sex chromosomes account for karyotype of 45,X[13]/46,X,+mar1[6]/46,X,+mar2[9], and approximately 0.5% of live births. Individuals with mosaic prenatal ultrasound was unremarkable. The woman underwent structural aberrations of the X and Y chromosomes exhibit repeat amniocentesis at 23 weeks and 4 days of gestation for complicated and variable phenotypes. The phenotypes of molecular detection. -

Background Briefing Document from the Consortium of Sponsors for The
BRIEFING BOOK FOR Pediatric Advisory Committee (PAC) Prepared by Alan Rogol, M.D., Ph.D. Prepared for Consortium of Sponsors Meeting Date: April 8th, 2019 TABLE OF CONTENTS LIST OF FIGURES ................................................... ERROR! BOOKMARK NOT DEFINED. LIST OF TABLES ...........................................................................................................................4 1. INTRODUCTION AND BACKGROUND FOR THE MEETING .............................6 1.1. INDICATION AND USAGE .......................................................................................6 2. SPONSOR CONSORTIUM PARTICIPANTS ............................................................6 2.1. TIMELINE FOR SPONSOR ENGAGEMENT FOR PEDIATRIC ADVISORY COMMITTEE (PAC): ............................................................................6 3. BACKGROUND AND RATIONALE .........................................................................7 3.1. INTRODUCTION ........................................................................................................7 3.2. PHYSICAL CHANGES OF PUBERTY ......................................................................7 3.2.1. Boys ..............................................................................................................................7 3.2.2. Growth and Pubertal Development ..............................................................................8 3.3. AGE AT ONSET OF PUBERTY.................................................................................9 3.4. -
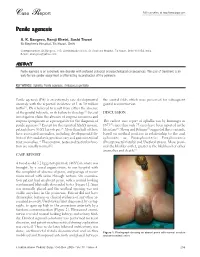
Case Report Full Text Online At
Case Report Full text online at http://www.jiaps.com Penile agenesis A. K. Bangroo, Ramji Khetri, Sashi Tiwari St Stephen's Hospital, Tis Hazari, Delhi Correspondence: AK Bangroo, 103, Administrative block, St. Stephens Hospital, Tis Hazari, Delhi-110054, India. E-mail: [email protected] ABSTRACT Penile agenesis is an extremely rare disorder with profound urological and psychological consequences. The goal of treatment is an early female gender assignment and feminizing reconstruction of the perineum. KEY WORDS: Aphallia, Penile agenesis, Ambiguous genitalia Penile agenesis (PA) is an extremely rare developmental the scrotal folds which were preserved for subsequent anomaly with the reported incidence of 1 in 30 million genital reconstruction. births[1]. PA is believed to result from either the absence of the genital tubercle, or its failure to develop.[2] Several DISCUSSION investigators claim the absence of corpora cavernosa and corpora spongiosum as a prerequisite for the diagnosis of The earliest case report of aphallia was by Imminger in penile agenesis.[3] Except for the reported XX-XY mosaic, 1853[2] since then only 75 cases have been reported in the patients have 46 XY karyotypes.[4] More than half of these literature[6]. Skoog and Belman[5] suggested three variants, have associated anomalies, including developmental de based on urethral position in relationship to the anal fects of the caudal axis, genitourinary and gastrointestinal sphincter, as: Postsphincteric; Presphincteric tract anomalies.[5] The scrotum, testes and testicular func (Prostatorectal fistula) and Urethral atresia. More proxi tion are usually normal[2]. mal the bladder outlet, greater is the likelihood of other anomalies and death.[5] CASE REPORT A two-day-old 3.2 kg genotypic male (46XY) neonate was brought, by a social organization, to our hospital with the complaint of absence of penis, and passage of meco nium mixed with urine through rectum. -

The Human Y Chromosome's Azoospermia Factor B (Azfb) Region
18 ORIGINAL ARTICLE J Med Genet: first published as 10.1136/jmg.40.1.18 on 1 January 2003. Downloaded from The human Y chromosome’s azoospermia factor b (AZFb) region: sequence, structure, and deletion analysis in infertile men A Ferlin, E Moro, A Rossi, B Dallapiccola, C Foresta ............................................................................................................................. J Med Genet 2003;40:18–24 See end of article for authors’ affiliations Microdeletions of the Y chromosome long arm are the most common mutations in infertile males, where ....................... they involve one or more “azoospermia factors” (AZFa, b, and c). Understanding of the AZF structure and gene content and mapping of the deletion breakpoints in infertile men are still incomplete. We Correspondence to: Professor C Foresta, have assembled a complete 4.3 Mb map of AZFb and surrounding regions by means of 38 BAC University of Padova, clones. The proximal part of AZFb consists of large repeated sequences organised in palindromes, but Department of Medical and most of it is single copy sequence. A number of known and novel genes and gene families map in this Surgical Sciences, Clinica interval, and most of them are testis specific or have testis specific transcripts. STS mapping allowed us Medica 3, Via Ospedale to identify four severely infertile subjects with a deletion in AZFb with similar breakpoints, therefore 105, 35128 Padova, Italy; [email protected] suggesting a common deletion mechanism. This deletion includes at least five single copy genes and two duplicated genes, but does not remove the historical AZFb candidate gene RBMY1. These data Revised version received suggest that other genes in AZFb may have important roles in spermatogenesis. -

EAU Pocket Guidelines on Male Hypogonadism 2013
GUIDELINES ON MALE HYPOGONADISM G.R. Dohle (chair), S. Arver, C. Bettocchi, S. Kliesch, M. Punab, W. de Ronde Introduction Male hypogonadism is a clinical syndrome caused by andro- gen deficiency. It may adversely affect multiple organ func- tions and quality of life. Androgens play a crucial role in the development and maintenance of male reproductive and sexual functions. Low levels of circulating androgens can cause disturbances in male sexual development, resulting in congenital abnormalities of the male reproductive tract. Later in life, this may cause reduced fertility, sexual dysfunc- tion, decreased muscle formation and bone mineralisation, disturbances of fat metabolism, and cognitive dysfunction. Testosterone levels decrease as a process of ageing: signs and symptoms caused by this decline can be considered a normal part of ageing. However, low testosterone levels are also associated with several chronic diseases, and sympto- matic patients may benefit from testosterone treatment. Androgen deficiency increases with age; an annual decline in circulating testosterone of 0.4-2.0% has been reported. In middle-aged men, the incidence was found to be 6%. It is more prevalent in older men, in men with obesity, those with co-morbidities, and in men with a poor health status. Aetiology and forms Male hypogonadism can be classified in 4 forms: 1. Primary forms caused by testicular insufficiency. 2. Secondary forms caused by hypothalamic-pituitary dysfunction. 164 Male Hypogonadism 3. Late onset hypogonadism. 4. Male hypogonadism due to androgen receptor insensitivity. The main causes of these different forms of hypogonadism are highlighted in Table 1. The type of hypogonadism has to be differentiated, as this has implications for patient evaluation and treatment and enables identification of patients with associated health problems. -

Targeting Lineage-Specific MITF Pathway in Human Melanoma Cell Lines by A-485, the Selective Small Molecule Inhibitor of P300/CBP
Author Manuscript Published OnlineFirst on September 28, 2018; DOI: 10.1158/1535-7163.MCT-18-0511 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Targeting lineage-specific MITF pathway in human melanoma cell lines by A-485, the selective small molecule inhibitor of p300/CBP Rui Wang1,2, Yupeng He1, Valerie Robinson1, Ziping Yang1, Paul Hessler1, Loren M. Lasko1, Xin Lu1, Anahita Bhathena1, Albert Lai1, Tamar Uziel1, Lloyd T. Lam1,3 1Oncology Discovery, AbbVie, 1 N Waukegan Road, North Chicago, IL 60064-6101, USA. 2Former AbbVie employee Running title: p300/CBP inhibition targets MITF in melanoma cell lines Keywords: p300/CBP inhibitor, melanoma, MITF, A-485, senescence Word count: 3,040 (excluding the abstract, references and legends) References: 29 3Correspondence: Lloyd T. Lam, Oncology Discovery, AP10-214, Dept. R4CD 1 North Waukegan Road North Chicago, IL 60064-6098, USA. Phone: 847-937-5585 Fax: 847-935-4994 E-mail: [email protected] Financial information Y.H., V.R., Z.Y., P.H., L.M.L., X.L, A.B., A.L., T.U., L.T.L. are employees of AbbVie. R.W. was an employee of AbbVie at the time of the study. The design, study conduct, and financial support for this research were provided by AbbVie. AbbVie participated in the interpretation of data, review, and approval of the manuscript. R.W., Y.H., V.R., Z.Y., P.H., L.M.L., X.L, A.B., A.L., T.U., L.T.L. have nothing to disclose. Downloaded from mct.aacrjournals.org on September 27, 2021. -

A MRI Diagnosis of Congenital Urogenital Anomalies in 27 Years
Journal of Advances in Radiology and Medical Imaging Volume 4 | Issue 1 ISSN: 2456-5504 Case Report Open Access A MRI Diagnosis of Congenital Urogenital Anomalies in 27 Years Old Man D’Amato D*, Ranalli T, Tatulli D, Bocchinfuso F, Manenti G, Valente F and Bizzaglia M Diagnostic and Interventional Radiology, Policlinico Tor Vergata, University of Rome “Tor Vergata”, Rome, Italy *Corresponding author: D’Amato D, Diagnostic and Interventional Radiology, Policlinico Tor Vergata, University of Rome “Tor Vergata”, Viale Oxford 181, Rome, Italy, Tel: +393207034690, E-mail: [email protected] Citation: D’Amato D, Ranalli T, Tatulli D, Bocchinfuso F, Manenti G, et al. (2019) A MRI Diagnosis of Congenital Urogenital Anomalies in 27 Years Old Man. J Adv Radiol Med Image 4(1): 102 Received Date: April 04, 2019 Accepted Date: August 26, 2019 Published Date: August 28, 2019 Abstract Congenital anorchia is an uncommon clinical condition. Etiology and pathogenetic mechanisms are often unknown. Although some patients with anorchia present with ambiguous external genitalia or micropenis, most have a normal phenotype. XY Disorders of Sex Development classifications are numerous and success rate in establishing a precise diagnosis is far lower than in XX karyotype. We report the case of a young man, with 46 XY karyotype showing various uro-genital abnormalities. A definitive diagnosis was not established due to the complex clinical presentation. Ultrasonography and Magnetic Resonance Imaging techniques were useful tools in the definition of uro-genital anomalies and gonadal development in this complex scenario. Keywords: Anorchia; Cryptorchidism; Urogenital Anomalies; DSD; MRI List of abbreviations: MRI: Magnetic Resonance Imaging; US: Ultrasonography; DSD: Disorders of Sex Development, FSH: Follicle- Stimulating Hormone; HCG: Human Chorionic Gonadotropin; LH: Luteinizing Hormone; AMH Antimüllerian Hormone; LHRH LH- Releasing Hormone; SD: Standard Deviation Introduction The disorders of sexual differentiation constitute a challenging area for both diagnostic and therapeutic impact. -

Program Nr: 1 from the 2004 ASHG Annual Meeting Mutations in A
Program Nr: 1 from the 2004 ASHG Annual Meeting Mutations in a novel member of the chromodomain gene family cause CHARGE syndrome. L.E.L.M. Vissers1, C.M.A. van Ravenswaaij1, R. Admiraal2, J.A. Hurst3, B.B.A. de Vries1, I.M. Janssen1, W.A. van der Vliet1, E.H.L.P.G. Huys1, P.J. de Jong4, B.C.J. Hamel1, E.F.P.M. Schoenmakers1, H.G. Brunner1, A. Geurts van Kessel1, J.A. Veltman1. 1) Dept Human Genetics, UMC Nijmegen, Nijmegen, Netherlands; 2) Dept Otorhinolaryngology, UMC Nijmegen, Nijmegen, Netherlands; 3) Dept Clinical Genetics, The Churchill Hospital, Oxford, United Kingdom; 4) Children's Hospital Oakland Research Institute, BACPAC Resources, Oakland, CA. CHARGE association denotes the non-random occurrence of ocular coloboma, heart defects, choanal atresia, retarded growth and development, genital hypoplasia, ear anomalies and deafness (OMIM #214800). Almost all patients with CHARGE association are sporadic and its cause was unknown. We and others hypothesized that CHARGE association is due to a genomic microdeletion or to a mutation in a gene affecting early embryonic development. In this study array- based comparative genomic hybridization (array CGH) was used to screen patients with CHARGE association for submicroscopic DNA copy number alterations. De novo overlapping microdeletions in 8q12 were identified in two patients on a genome-wide 1 Mb resolution BAC array. A 2.3 Mb region of deletion overlap was defined using a tiling resolution chromosome 8 microarray. Sequence analysis of genes residing within this critical region revealed mutations in the CHD7 gene in 10 of the 17 CHARGE patients without microdeletions, including 7 heterozygous stop-codon mutations. -

Taf7l Cooperates with Trf2 to Regulate Spermiogenesis
Taf7l cooperates with Trf2 to regulate spermiogenesis Haiying Zhoua,b, Ivan Grubisicb,c, Ke Zhengd,e, Ying Heb, P. Jeremy Wangd, Tommy Kaplanf, and Robert Tjiana,b,1 aHoward Hughes Medical Institute and bDepartment of Molecular and Cell Biology, Li Ka Shing Center for Biomedical and Health Sciences, California Institute for Regenerative Medicine Center of Excellence, University of California, Berkeley, CA 94720; cUniversity of California Berkeley–University of California San Francisco Graduate Program in Bioengineering, University of California, Berkeley, CA 94720; dDepartment of Animal Biology, University of Pennsylvania School of Veterinary Medicine, Philadelphia, PA 19104; eState Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 210029, People’s Republic of China; and fSchool of Computer Science and Engineering, The Hebrew University of Jerusalem, Jerusalem 91904, Israel Contributed by Robert Tjian, September 11, 2013 (sent for review August 20, 2013) TATA-binding protein (TBP)-associated factor 7l (Taf7l; a paralogue (Taf4b; a homolog of Taf4) (9), TBP-related factor 2 (Trf2) (10, of Taf7) and TBP-related factor 2 (Trf2) are components of the core 11), and Taf7l (12, 13). For example, mice bearing mutant or promoter complex required for gene/tissue-specific transcription deficient CREM showed decreased postmeiotic gene expression of protein-coding genes by RNA polymerase II. Previous studies and defective spermiogenesis (14). Mice deficient in Taf4b, reported that Taf7l knockout (KO) mice exhibit structurally abnor- a testis-specific homolog of Taf4, are initially normal but undergo mal sperm, reduced sperm count, weakened motility, and compro- progressive germ-cell loss and become infertile by 3 mo of age −/Y mised fertility. -

A Mutation in the Putative MLH3 Endonuclease Domain Confers a Defect in Both Mismatch Repair and Meiosis in Saccharomyces Cerevisiae
Copyright Ó 2008 by the Genetics Society of America DOI: 10.1534/genetics.108.086645 A Mutation in the Putative MLH3 Endonuclease Domain Confers a Defect in Both Mismatch Repair and Meiosis in Saccharomyces cerevisiae K. T. Nishant, Aaron J. Plys and Eric Alani1 Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York 14853-2703 Manuscript received January 2, 2008 Accepted for publication March 20, 2008 ABSTRACT Interference-dependent crossing over in yeast and mammalian meioses involves the mismatch repair protein homologs MSH4-MSH5 and MLH1-MLH3. The MLH3 protein contains a highly conserved metal- binding motif DQHA(X)2E(X)4E that is found in a subset of MLH proteins predicted to have endonuclease activities (Kadyrov et al. 2006). Mutations within this motif in human PMS2 and Saccharomyces cerevisiae PMS1 disrupted the endonuclease and mismatch repair activities of MLH1-PMS2 and MLH1-PMS1, re- spectively (Kadyrov et al. 2006, 2007; Erdeniz et al. 2007). As a first step in determining whether such an activity is required during meiosis, we made mutations in the MLH3 putative endonuclease domain motif (-D523N, -E529K) and found that single and double mutations conferred mlh3-null-like defects with respect to meiotic spore viability and crossing over. Yeast two-hybrid and chromatography analyses showed that the interaction between MLH1 and mlh3-D523N was maintained, suggesting that the mlh3-D523N mutation did not disrupt the stability of MLH3. The mlh3-D523N mutant also displayed a mutator phenotype in vegetative growth that was similar to mlh3D. Overexpression of this allele conferred a dominant-negative phenotype with respect to mismatch repair. -

Discovery of Candidate Genes for Stallion Fertility from the Horse Y Chromosome
DISCOVERY OF CANDIDATE GENES FOR STALLION FERTILITY FROM THE HORSE Y CHROMOSOME A Dissertation by NANDINA PARIA Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY August 2009 Major Subject: Biomedical Sciences DISCOVERY OF CANDIDATE GENES FOR STALLION FERTILITY FROM THE HORSE Y CHROMOSOME A Dissertation by NANDINA PARIA Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Approved by: Chair of Committee, Terje Raudsepp Committee Members, Bhanu P. Chowdhary William J. Murphy Paul B. Samollow Dickson D. Varner Head of Department, Evelyn Tiffany-Castiglioni August 2009 Major Subject: Biomedical Sciences iii ABSTRACT Discovery of Candidate Genes for Stallion Fertility from the Horse Y Chromosome. (August 2009) Nandina Paria, B.S., University of Calcutta; M.S., University of Calcutta Chair of Advisory Committee: Dr. Terje Raudsepp The genetic component of mammalian male fertility is complex and involves thousands of genes. The majority of these genes are distributed on autosomes and the X chromosome, while a small number are located on the Y chromosome. Human and mouse studies demonstrate that the most critical Y-linked male fertility genes are present in multiple copies, show testis-specific expression and are different between species. In the equine industry, where stallions are selected according to pedigrees and athletic abilities but not for reproductive performance, reduced fertility of many breeding stallions is a recognized problem. Therefore, the aim of the present research was to acquire comprehensive information about the organization of the horse Y chromosome (ECAY), identify Y-linked genes and investigate potential candidate genes regulating stallion fertility. -

Growth Hormone Deficiency Causing Micropenis:Peter A
Growth Hormone Deficiency Causing Micropenis:Peter A. Lee, MD, PhD, a Tom Mazur, PsyD, Lessons b Christopher P. Houk, MD, Learned c Robert M. Blizzard, MD d From a Well-Adjusted Adultabstract This report of a 46, XY patient born with a micropenis consistent with etiology from isolated congenital growth hormone deficiency is used to (1) raise the question regarding what degree testicular testosterone exposure aDepartment of Pediatrics, College of Medicine, Penn State to the central nervous system during fetal life and early infancy has on the University, Hershey, Pennsylvania; bCenter for Psychosexual development of male gender identity, regardless of gender of rearing; (2) Health, Jacobs School of Medicine and Biomedical suggest the obligatory nature of timely full disclosure of medical history; Sciences, University at Buffalo and John R. Oishei Children’s Hospital, Buffalo, New York; cDepartment of (3) emphasize that virtually all 46, XY infants with functional testes and Pediatrics, Medical College of Georgia, Augusta University, a micropenis should be initially boys except some with partial androgen Augusta, Georgia; and dDepartment of Pediatrics, College of Medicine, University of Virginia, Charlottesville, Virginia insensitivity syndrome; and (4) highlight the sustaining value of a positive long-term relationship with a trusted physician (R.M.B.). When this infant Dr Lee reviewed and discussed the extensive medical records with Dr Blizzard, reviewed presented, it was commonly considered inappropriate to gender assign an pertinent medical literature, and wrote each draft infant male whose penis was so small that an adult size was expected to be of the manuscript with input from all coauthors; inadequate, even if the karyotype was 46, XY, and testes were functional.