Speciation and Population Structure of Three Trichiurus Species Based
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hairtail and Frostfish (Trichiuridae) Exploitation Status Undefined
I & I NSW WILD FISHERIES RESEARCH PROGRAM Hairtail and Frostfish (Trichiuridae) EXPLOITATION STATUS UNDEFINED No local biological information available for either species in this group, but growth and maturity have been studied for Trichiurus lepturus from the East China Sea, where it supports a major fishery. SCIENTIFIC NAME STANDARD NAME COMMENT Trichiurus lepturus largehead hairtail Lepidopus caudatus frostfish Trichiurus lepturus Image © Bernard Yau Background The hairtail is commonly around 100 cm in length and about 2 kg in weight but reaches a The largehead hairtail (Trichiurus lepturus) maximum length of about 220 cm and weight belongs to the family Trichiuridae which, of 3.5 kg. worldwide, includes nine genera and about 30 species generally referred to as cutlassfishes or Overseas studies have observed that adults scabbardfishes. Off NSW, at least four species feed at the surface during the day, and retreat of trichiurids are found in deepwater, but the to deeper waters at night. In contrast, juveniles most well known member of the family to most and small adults tend to feed at night at the people is the hairtail, found in shallow coastal surface, and aggregate into schools at depths waters and estuaries. during the day. The adult hairtail diet consists mainly of fish with occasional squid and A cosmopolitan species, the largehead hairtail crustaceans, whereas juveniles mainly feed on is subject to significant fisheries off many planktonic crustaceans, euphausiids and small Asian countries, particularly China and Korea. fish. The world catch reportedly now exceeds 1.5 million t annually. In eastern Australia, Reported landings in NSW generally range hairtail occasionally school in coastal bays between 10 and 25 t with catches greatest and estuaries where they may be targeted during March-May. -

Fishes of Terengganu East Coast of Malay Peninsula, Malaysia Ii Iii
i Fishes of Terengganu East coast of Malay Peninsula, Malaysia ii iii Edited by Mizuki Matsunuma, Hiroyuki Motomura, Keiichi Matsuura, Noor Azhar M. Shazili and Mohd Azmi Ambak Photographed by Masatoshi Meguro and Mizuki Matsunuma iv Copy Right © 2011 by the National Museum of Nature and Science, Universiti Malaysia Terengganu and Kagoshima University Museum All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means without prior written permission from the publisher. Copyrights of the specimen photographs are held by the Kagoshima Uni- versity Museum. For bibliographic purposes this book should be cited as follows: Matsunuma, M., H. Motomura, K. Matsuura, N. A. M. Shazili and M. A. Ambak (eds.). 2011 (Nov.). Fishes of Terengganu – east coast of Malay Peninsula, Malaysia. National Museum of Nature and Science, Universiti Malaysia Terengganu and Kagoshima University Museum, ix + 251 pages. ISBN 978-4-87803-036-9 Corresponding editor: Hiroyuki Motomura (e-mail: [email protected]) v Preface Tropical seas in Southeast Asian countries are well known for their rich fish diversity found in various environments such as beautiful coral reefs, mud flats, sandy beaches, mangroves, and estuaries around river mouths. The South China Sea is a major water body containing a large and diverse fish fauna. However, many areas of the South China Sea, particularly in Malaysia and Vietnam, have been poorly studied in terms of fish taxonomy and diversity. Local fish scientists and students have frequently faced difficulty when try- ing to identify fishes in their home countries. During the International Training Program of the Japan Society for Promotion of Science (ITP of JSPS), two graduate students of Kagoshima University, Mr. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

Morphology and Phylogenetic Relationships of Fossil Snake Mackerels and Cutlassfishes (Trichiuroidea) from the Eocene (Ypresian) London Clay Formation
MS. HERMIONE BECKETT (Orcid ID : 0000-0003-4475-021X) DR. ZERINA JOHANSON (Orcid ID : 0000-0002-8444-6776) Article type : Original Article Handling Editor: Lionel Cavin Running head: Relationships of London Clay trichiuroids Hermione Becketta,b, Sam Gilesa, Zerina Johansonb and Matt Friedmana,c aDepartment of Earth Sciences, University of Oxford, South Parks Road, Oxford, OX1 3AN, UK bDepartment of Earth Sciences, Natural History Museum, London, SW7 5BD, UK cCurrent address: Museum of Paleontology and Department of Earth and Environmental Sciences, University of Michigan, 1109 Geddes Ave, Ann Arbor, MI 48109-1079, USA *Correspondence to: Hermione Beckett, +44 (0) 1865 272000 [email protected], Department of Earth Sciences, University of Oxford, Oxford, UK, OX1 3AN Short title: Relationships of London Clay trichiuroids Author Manuscript Key words: Trichiuroidea, morphology, London Clay, Trichiuridae, Gempylidae, fossil This is the author manuscript accepted for publication and has undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1002/spp2.1221 This article is protected by copyright. All rights reserved A ‘Gempylids’ (snake mackerels) and trichiurids (cutlassfishes) are pelagic fishes characterised by slender to eel-like bodies, deep-sea predatory ecologies, and large fang-like teeth. Several hypotheses of relationships between these groups have been proposed, but a consensus remains elusive. Fossils attributed to ‘gempylids’ and trichiurids consist almost exclusively of highly compressed body fossils and isolated teeth and otoliths. We use micro-computed tomography to redescribe two three- dimensional crania, historically assigned to †Eutrichiurides winkleri and †Progempylus edwardsi, as well as an isolated braincase (NHMUK PV OR 41318). -

Fao Species Catalogue
FAO Fisheries Synopsis No. 125, Volume 15 ISSN 0014-5602 FIR/S1 25 Vol. 15 FAO SPECIES CATALOGUE VOL. 15. SNAKE MACKERELS AND CUTLASSFISHES OF THE WORLD (FAMILIES GEMPYLIDAE AND TRICHIURIDAE) AN ANNOTATED AND ILLUSTRATED CATALOGUE OF THE SNAKE MACKERELS, SNOEKS, ESCOLARS, GEMFISHES, SACKFISHES, DOMINE, OILFISH, CUTLASSFISHES, SCABBARDFISHES, HAIRTAILS AND FROSTFISHES KNOWN TO DATE 12®lÄSÄötfSE, FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS FAO Fisheries Synopsis No. 125, Volume 15 FIR/S125 Vol. 15 FAO SPECIES CATALOGUE VOL. 15. SNAKE MACKERELS AND CUTLASSFISHES OF THE WORLD (Families Gempylidae and Trichiuridae) An Annotated and Illustrated Catalogue of the Snake Mackerels, Snoeks, Escolars, Gemfishes, Sackfishes, Domine, Oilfish, Cutlassfishes, Scabbardfishes, Hairtails, and Frostfishes Known to Date by I. Nakamura Fisheries Research Station Kyoto University Maizuru, Kyoto, 625, Japan and N. V. Parin P.P. Shirshov Institute of Oceanology Academy of Sciences Krasikova 23 Moscow 117218, Russian Federation FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS Rome, 1993 The designations employed and the presenta tion of material in this publication do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. M -40 ISBN 92-5-103124-X All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying or otherwise, without the prior permission of the copyright owner. Applications for such permission, with a statement of the purpose and extent of the reproduction, should be addressed to the Director, Publications Division, Food and Agriculture Organization of the United Nations, Via delle Terme di Caracalla, 00100 Rome, Italy. -

Fish in Disguise: Seafood Fraud in Korea
Fish in disguise: Seafood fraud in Korea A briefing by the Environmental Justice Foundation 1 Executive summary Between January and December 2018, the Environmental Justice Foundation (EJF) used DNA testing to determine levels of seafood fraud in the Republic of Korea. The results showed that over a third of samples tested were mislabelled. This mislabelling defrauds consumers, risks public health, harms the marine environment and can be associated with serious human rights abuses across the world. These findings demonstrate the urgent need for greater transparency and traceability in Korean seafood, including imported products. Key findings: • Over a third of seafood samples (34.8%, 105 of 302 samples) genetically analysed were mislabelled. • Samples labelled Fleshy Prawn, Fenneropenaeus chinensis (100%), Japanese Eel, Anguilla japonica (67.7%), Mottled Skate, Raja pulchra (53.3%) and Common Octopus, Octopus vulgaris (52.9%) had the highest rates of mislabelling. • Not a single sample labelled Fleshly Prawn was the correct species. • Mislabelling was higher in restaurants, fish markets and online than in general markets or superstores. • By processed types, sushi (53.9%), fresh fish (38.9%) and sashimi (33.6%) were the most likely to be mislabelled. • The seafood fraud identified by this research has direct negative impacts for consumers. It is clear that for some species sampled consumers were likely to be paying more than they should. For example, more than half of the eel and skate samples that were labelled domestic were actually found to be imported, which can cost only half of the price of domestic products. Swordfish mislabelled as Bluefin Tuna can be sold for four to five times as much. -

ILLEGAL FISHING Which Fish Species Are at Highest Risk from Illegal and Unreported Fishing?
ILLEGAL FISHING Which fish species are at highest risk from illegal and unreported fishing? October 2015 CONTENTS EXECUTIVE SUMMARY 3 INTRODUCTION 4 METHODOLOGY 5 OVERALL FINDINGS 9 NOTES ON ESTIMATES OF IUU FISHING 13 Tunas 13 Sharks 14 The Mediterranean 14 US Imports 15 CONCLUSION 16 CITATIONS 17 OCEAN BASIN PROFILES APPENDIX 1: IUU Estimates for Species Groups and Ocean Regions APPENDIX 2: Estimates of IUU Risk for FAO Assessed Stocks APPENDIX 3: FAO Ocean Area Boundary Descriptions APPENDIX 4: 2014 U.S. Edible Imports of Wild-Caught Products APPENDIX 5: Overexploited Stocks Categorized as High Risk – U.S. Imported Products Possibly Derived from Stocks EXECUTIVE SUMMARY New analysis by World Wildlife Fund (WWF) finds that over 85 percent of global fish stocks can be considered at significant risk of Illegal, Unreported, and Unregulated (IUU) fishing. This evaluation is based on the most recent comprehensive estimates of IUU fishing and includes the worlds’ major commercial stocks or species groups, such as all those that are regularly assessed by the United Nations Food and Agriculture Organization (FAO). Based on WWF’s findings, the majority of the stocks, 54 percent, are categorized as at high risk of IUU, with an additional 32 perent judged to be at moderate risk. Of the 567 stocks that were assessed, the findings show that 485 stocks fall into these two categories. More than half of the world’s most overexploited stocks are at the highest risk of IUU fishing. Examining IUU risk by location, the WWF analysis shows that in more than one-third of the world’s ocean basins as designated by the FAO, all of these stocks were at high or moderate risk of IUU fishing. -

1 Chapter 3.1.2: Longline Authors
3.1.2 LONGLINE CHAPTER 3.1.2: AUTHORS: LAST UPDATE: LONGLINE Domingo A., Forselledo R., Miller March 2014 P., Jiménez S., Mas F. and Pons M. 3.1.2 General description of longline fisheries 1. General description of longline gear and vessels Longline is a fishing gear that is comprised of a main line, with secondary lines attached and hooks placed on the end of them. Longline is a very old gear, derived from “volantín” or “three hook line”, which was used by the Phoenicians and the Egyptians in the Mediterranean Sea (Canterla, 1989). The word “longline” is possibly of Catalonian origin, and derived from southern Italian or Greece (Prat Sabater, 2006; González García, 2008). Since their beginnings and up to the 15th century at least, the longlines that were used were fixed, anchored to the coast with stalks. Later, in the open sea, “jeito” and “vareque” net gears were used which drifted while remaining moored to the vessels, and which could have given rise to drift gears, one of which is longline (Canterla, 1989). 1.a General description of pelagic longline 1.a.1 Introduction Drifting pelagic longline is used worldwide to catch widely distributed pelagic and semi-pelagic fish. This gear is very effective in catching tunas, billfish and sharks, among others (Doumenge, 1998; Matsuda, 1998). It consists of a main line or “mother” line, suspended in the water by secondary lines called float lines, which carry the floats. The branch lines hang from the main line and carry hooks on the ends. The characteristics of the materials, dimensions, types of floats and hooks, as well as the configuration of the lines are quite variable, depending mainly on the origin of the fleets, the fishermen and the target species. -

Intrinsic Vulnerability in the Global Fish Catch
The following appendix accompanies the article Intrinsic vulnerability in the global fish catch William W. L. Cheung1,*, Reg Watson1, Telmo Morato1,2, Tony J. Pitcher1, Daniel Pauly1 1Fisheries Centre, The University of British Columbia, Aquatic Ecosystems Research Laboratory (AERL), 2202 Main Mall, Vancouver, British Columbia V6T 1Z4, Canada 2Departamento de Oceanografia e Pescas, Universidade dos Açores, 9901-862 Horta, Portugal *Email: [email protected] Marine Ecology Progress Series 333:1–12 (2007) Appendix 1. Intrinsic vulnerability index of fish taxa represented in the global catch, based on the Sea Around Us database (www.seaaroundus.org) Taxonomic Intrinsic level Taxon Common name vulnerability Family Pristidae Sawfishes 88 Squatinidae Angel sharks 80 Anarhichadidae Wolffishes 78 Carcharhinidae Requiem sharks 77 Sphyrnidae Hammerhead, bonnethead, scoophead shark 77 Macrouridae Grenadiers or rattails 75 Rajidae Skates 72 Alepocephalidae Slickheads 71 Lophiidae Goosefishes 70 Torpedinidae Electric rays 68 Belonidae Needlefishes 67 Emmelichthyidae Rovers 66 Nototheniidae Cod icefishes 65 Ophidiidae Cusk-eels 65 Trachichthyidae Slimeheads 64 Channichthyidae Crocodile icefishes 63 Myliobatidae Eagle and manta rays 63 Squalidae Dogfish sharks 62 Congridae Conger and garden eels 60 Serranidae Sea basses: groupers and fairy basslets 60 Exocoetidae Flyingfishes 59 Malacanthidae Tilefishes 58 Scorpaenidae Scorpionfishes or rockfishes 58 Polynemidae Threadfins 56 Triakidae Houndsharks 56 Istiophoridae Billfishes 55 Petromyzontidae -

Authorship, Availability and Validity of Fish Names Described By
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Stuttgarter Beiträge Naturkunde Serie A [Biologie] Jahr/Year: 2008 Band/Volume: NS_1_A Autor(en)/Author(s): Fricke Ronald Artikel/Article: Authorship, availability and validity of fish names described by Peter (Pehr) Simon ForssSSkål and Johann ChrisStian FabricCiusS in the ‘Descriptiones animaliumÂ’ by CarsSten Nniebuhr in 1775 (Pisces) 1-76 Stuttgarter Beiträge zur Naturkunde A, Neue Serie 1: 1–76; Stuttgart, 30.IV.2008. 1 Authorship, availability and validity of fish names described by PETER (PEHR ) SIMON FOR ss KÅL and JOHANN CHRI S TIAN FABRI C IU S in the ‘Descriptiones animalium’ by CAR S TEN NIEBUHR in 1775 (Pisces) RONALD FRI C KE Abstract The work of PETER (PEHR ) SIMON FOR ss KÅL , which has greatly influenced Mediterranean, African and Indo-Pa- cific ichthyology, has been published posthumously by CAR S TEN NIEBUHR in 1775. FOR ss KÅL left small sheets with manuscript descriptions and names of various fish taxa, which were later compiled and edited by JOHANN CHRI S TIAN FABRI C IU S . Authorship, availability and validity of the fish names published by NIEBUHR (1775a) are examined and discussed in the present paper. Several subsequent authors used FOR ss KÅL ’s fish descriptions to interpret, redescribe or rename fish species. These include BROU ss ONET (1782), BONNATERRE (1788), GMELIN (1789), WALBAUM (1792), LA C E P ÈDE (1798–1803), BLO C H & SC HNEIDER (1801), GEO ff ROY SAINT -HILAIRE (1809, 1827), CUVIER (1819), RÜ pp ELL (1828–1830, 1835–1838), CUVIER & VALEN C IENNE S (1835), BLEEKER (1862), and KLUNZIN G ER (1871). -
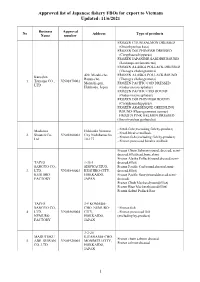
Approved List of Japanese Fishery Fbos for Export to Vietnam Updated: 11/6/2021
Approved list of Japanese fishery FBOs for export to Vietnam Updated: 11/6/2021 Business Approval No Address Type of products Name number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta) FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus) FROZEN JAPANESE SARDINE ROUND (Sardinops melanostictus) FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma) 420, Misaki-cho, FROZEN ALASKA POLLACK ROUND Kaneshin Rausu-cho, (Theragra chalcogramma) 1. Tsuyama CO., VN01870001 Menashi-gun, FROZEN PACIFIC COD DRESSED LTD Hokkaido, Japan (Gadus macrocephalus) FROZEN PACIFIC COD ROUND (Gadus macrocephalus) FROZEN DOLPHIN FISH ROUND (Coryphaena hippurus) FROZEN ARABESQUE GREENLING ROUND (Pleurogrammus azonus) FROZEN PINK SALMON DRESSED (Oncorhynchus gorbuscha) - Fresh fish (excluding fish by-product) Maekawa Hokkaido Nemuro - Fresh bivalve mollusk. 2. Shouten Co., VN01860002 City Nishihamacho - Frozen fish (excluding fish by-product) Ltd 10-177 - Frozen processed bivalve mollusk Frozen Chum Salmon (round, dressed, semi- dressed,fillet,head,bone,skin) Frozen Alaska Pollack(round,dressed,semi- TAIYO 1-35-1 dressed,fillet) SANGYO CO., SHOWACHUO, Frozen Pacific Cod(round,dressed,semi- 3. LTD. VN01840003 KUSHIRO-CITY, dressed,fillet) KUSHIRO HOKKAIDO, Frozen Pacific Saury(round,dressed,semi- FACTORY JAPAN dressed) Frozen Chub Mackerel(round,fillet) Frozen Blue Mackerel(round,fillet) Frozen Salted Pollack Roe TAIYO 3-9 KOMABA- SANGYO CO., CHO, NEMURO- - Frozen fish 4. LTD. VN01860004 CITY, - Frozen processed fish NEMURO HOKKAIDO, (excluding by-product) FACTORY JAPAN -

09 Kwok FISH 98(4)
748 Abstract—Age and growth of two spe- Age and growth of cutlassfi shes, Trichiurus spp., cies of cutlassfi shes, Trichiurus spp. (Trichiuridae), from the South China from the South China Sea Sea were examined. Between December 1996 and November 1997, 1495 speci- mens were collected from coastal waters Kai Yin Kwok near Hong Kong. Two species, Trichi- I-Hsun Ni urus lepturus and T. nanhaiensis, were Department of Biology harvested and ages of specimens were Hong Kong University of Science & Technology estimated by using transverse sections Hong Kong SAR, China of the sagittal otoliths. Opaque growth E-mail address (for I-H Ni, contact author): [email protected] rings were verifi ed to have formed annually during February. Lee’s phe- nomenon was not observed for either species, although T. lepturus tended to display reverse Lee’s phenomenon. Oto- lith weight was linearly related to age, The cutlassfi sh, Trichiurus lepturus China Sea suffer from overfi shing (Lin, and accounted for about 72% and 76% Linnaeus 1758, occurs throughout trop- 1985; Du et al., 1988; Ma and Xu, 1989; of the variation in age (t) for T. lepturus and T. nanhaiensis, respectively, com- ical and temperate waters of the world, Luo, 1991; Ye and Rosenberg, 1991; Xu parable to the von Bertalanffy growth between latitude 60°N and 45°S (Froese et al., 1994). It is harvested only in the models in preanal length (PL). For older and Pauly, 1997). World harvests are Bo Hai and the Yellow Sea as bycatch fi sh, otolith weight provided a more pre- approximately 750,000 tonnes annually in other fi sheries (Lin, 1985).