Transcriptomic Analysis Reveals Flavonoid Biosynthesis of Syringa Oblata Lindl
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Research Article Vein Distribution on the Deformation Behavior And
Hindawi Applied Bionics and Biomechanics Volume 2020, Article ID 8792143, 12 pages https://doi.org/10.1155/2020/8792143 Research Article Vein Distribution on the Deformation Behavior and Fracture Mechanisms of Typical Plant Leaves by Quasi In Situ Tensile Test under a Digital Microscope Jingjing Liu,1 Wei Ye ,1 Zhihui Zhang ,2 Zhenglei Yu,2 Hongyan Ding,1 Chao Zhang,1 and Sen Liu1 1Faculty of Mechanical & Material Engineering, Huaiyin Institute of Technology, Huai’an 223003, China 2The Key Laboratory of Engineering Bionic (Ministry of Education, China) and the College of Biological and Agricultural Engineering, Jilin University, 5988 Renmin Street, Changchun 130025, China Correspondence should be addressed to Wei Ye; [email protected] and Zhihui Zhang; [email protected] Received 12 December 2019; Revised 7 June 2020; Accepted 12 June 2020; Published 1 July 2020 Academic Editor: Raimondo Penta Copyright © 2020 Jingjing Liu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Angiosperm leaf venation is based on two major patterns, typically dicotyledonous branching and monocotyledonous parallel veins. The influence of these patterns on deformation and fracture properties is poorly understood. In this paper, three species of dicotyledons with netted venation and three species of monocots with parallel venation were selected, and the effect of vein distribution of leaves on their mechanical properties and deformation behavior was investigated. Whole images of leaves were captured using a digital camera, and their vein traits were measured using the image processing software Digimizer. -

FALL 2012 the Butterfly Garden Unfolds by THERESA ARIAL, BOTANIC GARDENS MANAGER
THE UNIVERSITY OF CALIFORNIA, RIVERSIDE BOTANIC GARDENS UCRBG Newsletter VOLUME 22, NUMBER 3 • FALL 2012 The Butterfly Garden Unfolds BY THERESA ARIAL, BOTANIC GARDENS MANAGER HE DEVELOPMENT OF any processes that have contributed to our adjusted the drainage swale to the garden, small and quaint to large unfolding garden. north end of the garden, blending Tand palatial, is a process. An idea is the Initially, we were very fortunate to form and function by incorporating begin ning of that process, and we have have a flat site for the Butterfly rocks specified by the original design. Ann Platzer to thank for that. Her Garden. With help from UCR’s To provide delineation and a form vision of devel oping a butterfly garden Agricultural Operations, we graded the that allowed the Passiflora to climb, we here at the UCRBG was the beginning area just before rains moved through. installed a four foot fence, running the of some thing beautiful. In fact, her This gave us the perfect opportunity to length of west side. Ten yards of idea was my inspiration to create a observe water movement, and make amend ment were delivered and incor - butterfly garden that would unfold any adjust ments. In the porated into the existing soil at a depth before our eyes. winter of 2010, we There are many factors to be consider ed when planning a garden. Among these are accessibility, design, plant selection, grading, drainage, and irrigation. To achieve our end goal has taken time and patience. The garden has been plant - ed since early May of 2012. -

CHARACTERISTICS of BLOOMING and POLLEN in FLOWERS of TWO Syringa SPECIES (F
ACTA AGROBOTANICA Vol. 66 (4), 2013: 65–72 Original Research Paper DOI: 10.5586/aa.2013.052 CHARACTERISTICS OF BLOOMING AND POLLEN IN FLOWERS OF TWO Syringa SPECIES (f. Oleaceae) Bożena Denisow, Monika Strzałkowska-Abramek Department of Botany, Laboratory of Horticultural Plant Biology, University of Life Sciences in Lublin Akademicka 15, 20-950 Lublin, Poland e-mail: [email protected] Received: 12.06.2013 Abstract Asia and within the range of their origin are consid- The observations were conducted in long-term sequen- ered as insect pollinated [2]. Lilacs, due to delicious ce studies in the years 2006, 2009, 2013, in the Lublin area, fragrant and abundant blooming, are one of the most Poland (51o 16’ N, 22o 30’ E). The flowering phenology, diurnal widely cultivated ornamental trees and shrubs in tem- pattern of blooming, pollen production and insect visitation to perate regions of the world [3]. Several hundred culti- the shrubs of Syringa oblata Lindl. var. dilatata (Nakai) Rehd. vars have been developed through extensive hybridiza- and S. meyeri ‘Palibin’ Schn. were examined. tion and artificial selection [4]. The plants (e.g. Syrin- Syringa oblata var. dilatata and S. meyeri ‘Palibin’ blo- ga vulgaris) have been used ethnobotanically to reduce omed from the mid May till mid June. The species are characte- fever and treat malaria, as a perfume and a tonic, and ristic of early morning diurnal pattern of blooming, with approx. in homeopathy [5]. of 60% of daily instalment of flowers opened before 9.00 (GMT The flowers are tiny, from the basic single, and + 2h). -

4. SYRINGA Linnaeus, Sp. Pl. 1: 9. 1753. 丁香 属 Ding Xiang Shu Shrubs Or Small Trees, Deciduous
Flora of China 15: 280–286. 1996. 4. SYRINGA Linnaeus, Sp. Pl. 1: 9. 1753. 丁香 属 ding xiang shu Shrubs or small trees, deciduous. Branchlets terete or 4-angled; pith solid; winter buds scaly, terminal buds often absent. Leaves opposite, simple or rarely pinnate, petiolate; leaf blade entire, pinnatisect or occasionally lobed. Inflorescences paniculate, terminal or lateral, generally composed of small cymes. Flowers bisexual, sessile or pedicellate. Calyx campanulate, regularly or irregularly 4-toothed or subtruncate, persistent. Corolla funnelform, salverform, or rotate; lobes 4, spreading or upright, valvate, usually cucullate and beaked at apex. Stamens 2, included or exserted. Ovules 2 in each locule, pendulous. Style filiform, shorter than stamens; stigma 2-cleft. Fruit a loculicidal capsule, slightly compressed. Seeds 2 in each locule, flat, narrowly winged; endosperm present; radicle erect. About 20 species: Afghanistan, India, Japan, Kashmir, Korea, Nepal, Pakistan, Sikkim; SW Asia, SE Europe; 16 species in China. Most species of Syringa are cultivated as ornamental plants, a few are medicinal. 1a. Corolla tube as long as calyx or slightly longer; filament exserted from corolla tube; corolla white or cream colored ............................................................................................. 16. S. reticulata 1b. Corolla tube 2.5–8 × longer than calyx; anthers wholly or partly included, occasionally exserted. 2a. Panicles terminal, or terminal and lateral. 3a. Anthers entirely exserted from corolla tube; flowers white ................................ 1. S. tibetica 3b. Anthers wholly or partly included in corolla tube; flowers red, purple, lilac, pink, or sometimes white. 4a. Leaf blade abaxially glabrous ................................................................. 2. S. yunnanensis 4b. Leaf blade abaxially ± hairy. 5a. Corolla tube funnelform, gradually enlarged above middle; corolla lobes usually upright. -

Leaf Fresh Weight Versus Dry Weight: Which Is Better for Describing the Scaling Relationship Between Leaf Biomass and Leaf Area for Broad-Leaved Plants?
Article Leaf Fresh Weight Versus Dry Weight: Which is Better for Describing the Scaling Relationship between Leaf Biomass and Leaf Area for Broad-Leaved Plants? Weiwei Huang 1, David A. Ratkowsky 2 , Cang Hui 3, Ping Wang 1, Jialu Su 1 and Peijian Shi 1,* 1 Co-Innovation Centre for Sustainable Forestry in Southern China, Bamboo Research Institute, College of Biology and the Environment, Nanjing Forestry University, 159 Longpan Road, Nanjing 210037, China; [email protected] (W.H.); [email protected] (P.W.); [email protected] (J.S.) 2 Tasmanian Institute of Agriculture, University of Tasmania, Private Bag 98, Hobart, TAS 7001, Australia; [email protected] 3 Centre for Invasion Biology, Department of Mathematical Sciences, Stellenbosch University, Matieland 7602, South Africa; [email protected] * Correspondence: [email protected]; Tel.: +86-25-85427231 Received: 9 February 2019; Accepted: 11 March 2019; Published: 13 March 2019 Abstract: Leaf dry mass per unit area (LMA) is considered to represent the photosynthetic capacity, which actually implies a hypothesis that foliar water mass (leaf fresh weight minus leaf dry weight) is proportional to leaf dry weight during leaf growth. However, relevant studies demonstrated that foliar water mass disproportionately increases with increasing leaf dry weight. Although scaling relationships of leaf dry weight vs. leaf area for many plants were investigated, few studies compared the scaling relationship based on leaf dry weight with that based on leaf fresh weight. In this study, we used the data of three families (Lauraceae, Oleaceae, and Poaceae, subfamily Bambusoideae) with five broad-leaved species for each family to examine whether using different measures for leaf biomass (i.e., dry weight and fresh weight) can result in different fitted results for the scaling relationship between leaf biomass and area. -

Lilac Newsletter Vol
Lilac Newsletter Vol. IX, No.4, April, 1983 INTERNATIONAL LILAC SOCIETY INTERNA TlONAL LILAC SOCIETY is a non-profit corporation comprised of individuals who share a particular interest, appre- ciation and fondness for lilacs. Through exchange of knowledge, experience and facts gained by members it is helping to promote, educate and broaden public understanding and awareness. Articles printed in this publication are the views and opinions of the authorts) and do not necessarily represent those of the editor or the International Lilac Society. ' This publication, LILAC NEWSLETTER (formerly THE PIPELINE) is issued monthly. Back copies are available by writing to the Internationallilac Society, c/o Mr. Charles Holetich, Royal Botanical Gardens, Box 399, Hamilton, Ontario, Canada. L8N 3H8. Please send 50 cents for each copy requested. President: Dr. Owen M. Rogers, University of New Hampshire, Dept. of Plant Science, Nesmith Hall, Durham, NH 03824 Secretary: Walter W. Oakes" Box 315, Rumford, Maine, 04276 Treasurer: Mrs. Marie Chaykowski 4041 Winchell Road, Mantua, Ohio 44255 Editor: Ms. Mary C. Smith, Hte. 2, Bellevue, Iowa 52031 INTERNA nONAL LILAC SOCIETY, William A. Utley, Ex. Vice-Pres., Grape Hill Farm, Devereaux Rd., Clyde, NY 14433 MEMBERSHIP CLASSIFICA nON Single annual $ 7.50 " Family . 10.00 Sustaining . 15.00 Institutional iCommercial . 20.00 ". life. " . 150.00 "Mail membership dues to I. L.S. Secretary -1- LILACS by Judith Hillstrom* **Reprint Does growing a "lilac tree" 12ique your sense of garden adventure? This is exactly what I intend. The iinage a 1I1ilac tree" presents is one of whimsy and artistry hand in hand - a smooth but graceful trunk topped by a rounded crown of lacy plumes to scent the spring breeze. -

Sewage Purification Effects of Several Ornamental Shrubs
2018 7th International Conference on Social Science, Education and Humanities Research (SSEHR 2018) Sewage Purification Effects of Several Ornamental Shrubs Bo Yang1,2, Jie Tang1 1College of Environment and Resource, Jilin University, Changchun, Jilin Province, China. 2College of Agronomy, Jilin Agricultural Science and Technology University, Jilin, Jilin Province, China. Keywords: shrubs; TP; TN; COD; purification rate. Abstract: Using the simulated planting method, this paper compares the sewage purification effect of 7 kinds of ornamental shrubs on TP, TN and COD. The results show that, all shrubs have obvious purification effects on TP, TN and COD in sewage. The TP purification abilities of shrubs go as following: Spiraea japonica Gold Mound. > Amorpha fruticosa Linn. > Swida alba Opiz. > Ligustrum obtusifolium Sieb. > Lespedeza bicolor Turcz. > Syringa oblata Lindl. > Forsythio mandshurica Uyeki.. TN purification abilities of shrubs go as following: Amorpha fruticosa Linn. > Spiraea japonica Gold Mound. > Lespedeza bicolor Turcz. > Ligustrum obtusifolium Sieb. > Swida alba Opiz. > Forsythio mandshurica Uyeki. > Syringa oblata Lindl. COD purification abilities of shrubs go as following: Spiraea japonica Gold Mound. > Swida alba Opiz. > Syringa oblata Lindl. > Amorpha fruticosa Linn. > Lespedeza bicolor Turcz. > Ligustrum obtusifolium Sieb. > Forsythio mandshurica Uyeki. The purification rates show a declining trend then keep constant with time passing by. 1. Introduction To adopt plants in ecological restoration means to give full play to plants’ ecological benefits, and create conditions for the harmonious existence of human beings and water body by utilizing plants’ functions of soil and water conservation, erosion prevention and control, water purification, filtering and buffering, climate regulation, environmental beautification and biodiversity improvement. Previous studies on plant ecological restoration focused on aquatic plants. -

Breeze\Lilacs.Wpd
GROWING LILACS 100 East Center Street, L600 Provo, UT 84606 801-851-8460 Lilacs are versatile flowering shrubs which TYPES OF LILACS have a wide range of uses in the home garden. They can serve as border plants with There are many species and kinds of lilacs. smaller shrubs, as corner plantings, as Extensive cross-breeding, however, has windscreens, or as flowering hedges. Both made these species very much alike. Even plants and flowers are very attractive. botanical experts sometimes find them difficult to identify. Although lilacs display flowers that are among the most delicate of the ornamentals, All lilacs belong to the genus Syringa. the plants are among the most hardy. Some Common to most kinds are unlobed leaves varieties can survive winter temperatures of and flowers that grow in clusters. Among –60° F. They are therefore suited to all parts the best known are the following: of the United States except the South, where winters are too mild to provide the plants Common lilac (Syringa vulgaris), as the with the seasonal rest period they need. name implies, is the best known of all the lilacs in the United States. This shrub can be Lilac plants often grow and as tall as 20 feet, and the flowers are continue to flower for many fragrant and usually lilac-colored, although years even if totally they can be of other hues. Leaves are neglected. Normally, the somewhat heart-shaped and smooth. only care they need is pruning to keep them within Persian lilac (Syringa persica) can grow to a bounds. height of 10 feet. -
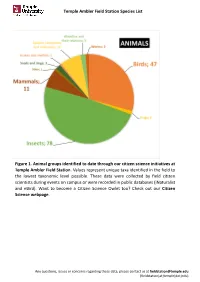
Temple Ambler Field Station Species List Figure 1. Animal Groups Identified to Date Through Our Citizen Science Initiatives at T
Temple Ambler Field Station Species List Figure 1. Animal groups identified to date through our citizen science initiatives at Temple Ambler Field Station. Values represent unique taxa identified in the field to the lowest taxonomic level possible. These data were collected by field citizen scientists during events on campus or were recorded in public databases (iNaturalist and eBird). Want to become a Citizen Science Owlet too? Check out our Citizen Science webpage. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List Figure 2. Plant diversity identified to date in the natural environments and designed gardens of the Temple Ambler Field Station and Ambler Arboretum. These values represent unique taxa identified to the lowest taxonomic level possible. Highlighted are 14 of the 116 flowering plant families present that include 524 taxonomic groups. A full list can be found in our species database. Cultivated specimens in our Greenhouse were not included here. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List database_title Temple Ambler Field Station Species List last_update 22October2020 description This database includes all species identified to their lowest taxonomic level possible in the natural environments and designed gardens on the Temple Ambler campus. These are occurrence records and each taxon is only entered once. This is an occurrence record, not an abundance record. IDs were performed by senior scientists and specialists, as well as citizen scientists visiting campus. -

Higher Plants Part A1
APPLICATION FOR CONSENT TO RELEASE A GMO – HIGHER PLANTS PART A1: INFORMATION REQUIRED UNDER SCHEDULE 1 OF THE GENETICALY MODIFIED ORGANISMS (DELIBERATE RELEASE) REGULATIONS 2002 PART 1 General information 1. The name and address of the applicant and the name, qualifications and experience of the scientist and of every other person who will be responsible for planning and carrying out the release of the organisms and for the supervision, monitoring and safety of the release. Applicant: Rothamsted Research, West Common, Harpenden Hertfordshire, AL5 2JQ UK 2. The title of the project. Study of aphid, predator and parasitoid behaviour in wheat producing aphid alarm pheromone PART II Information relating to the parental or recipient plant 3. The full name of the plant - (a) family name, Poaceae (b) genus, Triticum (c) species, aestivum (d) subspecies, N/A (e) cultivar/breeding line, Cadenza (f) common name. Common wheat/ bread wheat 4. Information concerning - (a) the reproduction of the plant: (i) the mode or modes of reproduction, (ii) any specific factors affecting reproduction, (iii) generation time; and (b) the sexual compatibility of the plant with other cultivated or wild plant species, including the distribution in Europe of the compatible species. ai) Reproduction is sexual leading to formation of seeds. Wheat is approximately 99% autogamous under natural field conditions; with self-fertilization normally occurring before flowers open. Wheat pollen grains are relatively heavy and any that are released from the flower remain viable for between a few minutes and a few hours. Warm, dry, windy conditions may increase cross- pollination rates on a variety to variety basis (see also 6 below). -

Diversity and Distribution of Floral Scent
The Botanical Review 72(1): 1-120 Diversity and Distribution of Floral Scent JETTE T. KNUDSEN Department of Ecology Lund University SE 223 62 Lund, Sweden ROGER ERIKSSON Botanical Institute GOteborg University Box 461, SE 405 30 GOteborg, Sweden JONATHAN GERSHENZON Max Planck Institute for Chemical Ecology Hans-KnOll Strasse 8, 07745 ,lena, Germany AND BERTIL ST,~HL Gotland University SE-621 67 Visby, Sweden Abstract ............................................................... 2 Introduction ............................................................ 2 Collection Methods and Materials ........................................... 4 Chemical Classification ................................................... 5 Plant Names and Classification .............................................. 6 Floral Scent at Different Taxonomic Levels .................................... 6 Population-Level Variation .............................................. 6 Species- and Genus-Level Variation ....................................... 6 Family- and Order-Level Variation ........................................ 6 Floral Scent and Pollination Biology ......................................... 9 Floral Scent Chemistry and Biochemistry ...................................... 10 Floral Scent and Evolution ................................................. 11 Floral Scent and Phylogeny ................................................ 12 Acknowledgments ....................................................... 13 Literature Cited ........................................................ -

Lilac Newsletter Vol
Lilac Newsletter Vol. VI, No.4, April, 1980. INTERNATIONAL LILAC SOCIETY INTERNATIONAL LILAC SOCIETY is a non-profit corporation comprised of individuals who share a particular interest, appre- ciation and fondness for lilacs. Through exchange of knowledge, experience and facts gained by members it is helping to promote, educate and broaden public understanding and awareness. Articles printed in this publication are the views and opinions of the authorls) and do not necessarily represent those of the editor or the International Lilac Society. This publication, LILAC NEWSLETTER (for- merly THE PIPELINE) is issued monthly. Back copies are available by writing to the Inter- national Lilac Society, c/o Mr. Charles Holetich, Royal Botanical Gardens, Box 399, Hamilton, Ontario, Canada. L8N 3H8. Please send 50 cents for each copy requested. President: Dr. Owen M. Rogers, University of New Hampshire, Dept. of Plant Science, Nesmith Hall, Durham, N.H. 03824. Secretary: Walter W. Oakes" Box 315, Rumford, Maine, 04276 Treasurer: Mrs. Marie Chaykowski 4041 Winchell Road, Mantua, Ohio, 44255 Editor: Walter E. Eickhorst 129 West Franklin St .• Naperville, Illinois. 60540 INTERNATIONAL LILAC SOCIETY, William A. Utley, Ex. Vice-Pres., Grape Hill Farm, Devereaux Rd., Clyde, NY 14433. MEMBERSHIP CLASSIFICATION 1980 ($ US) Single annual . 7.50 Family . 10.00 Sustaining . 15.00 Institutional/Commercial . 20.00 Life . 150.00 *Mail membership dues to 1.1.S. Secretary. -1- Feprint* LIIJ'CSFIU1 EARLYTIILLATE It HAS BEEN SAID THAT THE LILAC HOLDS A DEEPER FOOTHOLD IN man's AFFECTIM THAN ALRROST ANY shrub, AND THAT IN NO OTHER PLANT IS THE SPIRIT OF HONE RRORE INTIMATELY entwined.