Genomics of the Parasitic Nematode Ascaris and Its Relatives
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pulmonary Edema Associated with Ascaris Lumbricoides in a Patient with Mild Mitral Stenosis: a Case Report
Eur J Gen Med 2004; 1(2): 43-45 CASE REPORT PULMONARY EDEMA ASSOCIATED WITH ASCARIS LUMBRICOIDES IN A PATIENT WITH MILD MITRAL STENOSIS: A CASE REPORT Talantbek Batyraliev1, Beyhan Eryonucu2, Zarema Karben1, Hakan Sengul1, Niyazi Güler2, Orhan Dogru1, Alper Sercelik1 Sani Konukoglu Medical Center , Department of Cardiology1 Yüzüncü Yıl University, Department of Cardiology2 Ascaris lumbricoides remains the most common intestinal nematode in the world. Clinical manifestations of ascaris lumbricoides are different in each stage of the infection. We presented an unusual presentation of ascaris lumbricoides. Key words: Pulmonary oedema, mild mitral stenosis, Ascaris lumbricoides INTRODUCTION oedema due to mitral stenosis. Treatment of Ascaris lumbricoides (AL) remain the most diuretics was initiated. She was placed on common intestinal nematode in the world. oxygen by nasal cannula. Tachycardia was not Clinical manifestations of AL are different taken under control by treatment with digoxin in each stage of the infection. Infection with and verapamil. A reason for excessive nausea ascaris appears to be asymptomatic in the vast and vomiting was not determined and these majority of cases, but may produce serious semptoms were not responsive to antiemetic pulmonary disease or obstruction of biliary drugs. Despite intensive treatment, clinical or intestinal tract in a small proportion of improvement was not occured. Fortunately, at infected people. We presented an unusual the 3rd day, the patient was expelled ascaris presentation of AL and pulmonary edema lumbricoides (AL) (Figure). Once the AL was (1,2). removed the patient’s respiratory condition dramatically improved. The patient was CASE started on a 3-day course of mebendazole A 27-year-old woman was admitted to and discharged 4 days later in good general our hospital because of an episode of acute condition. -

The Functional Parasitic Worm Secretome: Mapping the Place of Onchocerca Volvulus Excretory Secretory Products
pathogens Review The Functional Parasitic Worm Secretome: Mapping the Place of Onchocerca volvulus Excretory Secretory Products Luc Vanhamme 1,*, Jacob Souopgui 1 , Stephen Ghogomu 2 and Ferdinand Ngale Njume 1,2 1 Department of Molecular Biology, Institute of Biology and Molecular Medicine, IBMM, Université Libre de Bruxelles, Rue des Professeurs Jeener et Brachet 12, 6041 Gosselies, Belgium; [email protected] (J.S.); [email protected] (F.N.N.) 2 Molecular and Cell Biology Laboratory, Biotechnology Unit, University of Buea, Buea P.O Box 63, Cameroon; [email protected] * Correspondence: [email protected] Received: 28 October 2020; Accepted: 18 November 2020; Published: 23 November 2020 Abstract: Nematodes constitute a very successful phylum, especially in terms of parasitism. Inside their mammalian hosts, parasitic nematodes mainly dwell in the digestive tract (geohelminths) or in the vascular system (filariae). One of their main characteristics is their long sojourn inside the body where they are accessible to the immune system. Several strategies are used by parasites in order to counteract the immune attacks. One of them is the expression of molecules interfering with the function of the immune system. Excretory-secretory products (ESPs) pertain to this category. This is, however, not their only biological function, as they seem also involved in other mechanisms such as pathogenicity or parasitic cycle (molting, for example). Wewill mainly focus on filariae ESPs with an emphasis on data available regarding Onchocerca volvulus, but we will also refer to a few relevant/illustrative examples related to other worm categories when necessary (geohelminth nematodes, trematodes or cestodes). -

Baylisascariasis
Baylisascariasis Importance Baylisascaris procyonis, an intestinal nematode of raccoons, can cause severe neurological and ocular signs when its larvae migrate in humans, other mammals and birds. Although clinical cases seem to be rare in people, most reported cases have been Last Updated: December 2013 serious and difficult to treat. Severe disease has also been reported in other mammals and birds. Other species of Baylisascaris, particularly B. melis of European badgers and B. columnaris of skunks, can also cause neural and ocular larva migrans in animals, and are potential human pathogens. Etiology Baylisascariasis is caused by intestinal nematodes (family Ascarididae) in the genus Baylisascaris. The three most pathogenic species are Baylisascaris procyonis, B. melis and B. columnaris. The larvae of these three species can cause extensive damage in intermediate/paratenic hosts: they migrate extensively, continue to grow considerably within these hosts, and sometimes invade the CNS or the eye. Their larvae are very similar in appearance, which can make it very difficult to identify the causative agent in some clinical cases. Other species of Baylisascaris including B. transfuga, B. devos, B. schroeder and B. tasmaniensis may also cause larva migrans. In general, the latter organisms are smaller and tend to invade the muscles, intestines and mesentery; however, B. transfuga has been shown to cause ocular and neural larva migrans in some animals. Species Affected Raccoons (Procyon lotor) are usually the definitive hosts for B. procyonis. Other species known to serve as definitive hosts include dogs (which can be both definitive and intermediate hosts) and kinkajous. Coatimundis and ringtails, which are closely related to kinkajous, might also be able to harbor B. -

Top Papers in Neglected Tropical Diseases Diagnosis
Top papers in Neglected Tropical Diseases Diagnosis Professor Peter L Chiodini ESCMID eLibrary © by author Neglected tropical diseases http://www.who.int/neglected_diseases/diseases/en/ Accessed 9th April 2017 • Buruli ulcer • Lymphatic filariasis • Chagas disease • Mycetoma • Dengue and Chikungunya • Onchocerciasis • Dracunculiasis • Rabies • Echinococcosis • Schistosomiasis • Foodborne • Soil-transmitted trematodiases helminthiases • Human African • Taeniasis/Cysticercosis trypanosomiasis • Trachoma • Leishmaniasis • Yaws • ESCMIDLeprosy eLibrary © by author PLOS Neglected Tropical Diseases http://journals.plos.org/plosntds/s/journal-information#loc-scope Accessed 9th April 2017 The “big three” diseases, HIV/AIDS, malaria, and tuberculosis, are not generally considered for PLOS Neglected Tropical Diseases. Plasmodium vivax possibly ESCMID eLibrary © by author Flynn, FV (1985) “As is your pathology, so is your practice” ESCMID eLibrary © by author Time for a Model List of Essential Diagnostics Schroeder LF et al. NEJM 2016; 374: 2511 ESCMID eLibrary © by author Peruvian Leishmania: braziliensis or mexicana? ESCMID eLibrary © by author Leishmania (Viannia) from Peru ESCMID eLibrary © by author Karimkhani C et al. Lancet Infect Dis 2016; 16: 584-591 • Global Burden of Disease Study “Unlike the incidence of most other neglected tropical diseases, the incidence of cutaneous leishmaniasis is increasing” ESCMID eLibrary © by author Karimkhani C et al. Lancet Infect Dis 2016; 16: 584-591 ESCMID eLibrary © by author Akhoundi M et al. Molecular Aspects of Medicine (2017) doi: 10.1016/ j.mam.2016.11.012. ESCMID eLibrary © by author Akhoundi M et al. Molecular Aspects of Medicine (2017) doi: 10.1016/ j.mam.2016.11.012. ESCMID eLibrary © by author Akhoundi M et al. Molecular Aspects of Medicine (2017) doi: 10.1016/ j.mam.2016.11.012. -

Hybrid Ascaris Suum/Lumbricoides (Ascarididae) Infestation in a Pig
Cent Eur J Public Health 2013; 21 (4): 224–226 HYBRID ASCARIS SUUM/LUMBRICOIDES (ASCARIDIDAE) INFESTATION IN A PIG FARMER: A RARE CASE OF ZOONOTIC ASCARIASIS Moreno Dutto1, Nicola Petrosillo2 1Department of Prevention, Local Health Unit, ASL CN1, Saluzzo (CN), Italy 2National Institute for Infectious Diseases “Lazzaro Spallanzani”, Rome, Italy SUMMARY We present a case of the 42 year old pig farmer from the province of Cuneo in Northwest Italy who was infected by the soil-transmitted nema- tode Ascaris sp. In November 2010 the patient found one worm in his stool, subsequently identified as female specimen of Ascaris sp. After a first anthelmintic treatment, another worm was found in his stool, that was later identified as male Ascaris sp. Blood tests prescribed by the patient’s family physician, as suggested by a parasitologist, found nothing abnormal. A chest x-ray was negative for Loeffler’s syndrome and an ultrasound of the abdomen was normal with no evidence of hepatic problems. The nematode collected from the patient was genetically characterized using the ribosomal nuclear marker ITS. The PCR-RFLP analysis showed a hybrid genotype, intermediate between A. suum/lumbricoides. It was subsequently ascertained that some pigs on the patient’s farm had A. suum infection; no other family member was infected. A cross- infestation from the pigs as source was the likely way of transmission. This conclusion is further warranted by the fact, that the patient is a confirmed nail-biter, a habit which facilitates oral-fecal transmission of parasites and pathogens. Key words: ascariasis, Ascaris suum, cross infection, pigs, zoonosis, hybrid genotype Address for correspondence: M. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Parascaris Univalens After in Vitro Exposure to Ivermectin, Pyrantel Citrate and Thiabendazole
Transcriptional responses in Parascaris univalens after in vitro exposure to ivermectin, pyrantel citrate and thiabendazole Frida Martin ( [email protected] ) Swedish University of Agricultural Sciences https://orcid.org/0000-0002-3149-3835 Faruk Dube Sveriges Lantbruksuniversitet Veterinarmedicin och husdjursvetenskap Oskar Karlsson Lindsjö Sveriges Lantbruksuniversitet Veterinarmedicin och husdjursvetenskap Matthías Eydal Haskoli Islands Johan Höglund Sveriges Lantbruksuniversitet Veterinarmedicin och husdjursvetenskap Tomas F. Bergström Sveriges Lantbruksuniversitet Veterinarmedicin och husdjursvetenskap Eva Tydén Sveriges Lantbruksuniversitet Veterinarmedicin och husdjursvetenskap Research Keywords: transcriptome, anthelmintic resistance, RNA sequencing, differential expression, lgc-37 Posted Date: March 18th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-17857/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published on July 9th, 2020. See the published version at https://doi.org/10.1186/s13071-020-04212-0. Page 1/23 Abstract Background: Parascaris univalens is a pathogenic parasite of foals and yearlings worldwide. In recent years Parascaris spp. worms have developed resistance to several of the commonly used anthelmintics, though currently the mechanisms behind this development is unknown. The aim of this study was to investigate the transcriptional responses in adult P. univalens worms after in vitro exposure -

High Heritability for Ascaris and Trichuris Infection Levels in Pigs
Heredity (2009) 102, 357–364 & 2009 Macmillan Publishers Limited All rights reserved 0018-067X/09 $32.00 www.nature.com/hdy ORIGINAL ARTICLE High heritability for Ascaris and Trichuris infection levels in pigs P Nejsum1,2, A Roepstorff1,CBJrgensen2, M Fredholm2, HHH Go¨ring3, TJC Anderson3 and SM Thamsborg1 1Danish Centre for Experimental Parasitology, Department of Veterinary Pathobiology, Faculty of Life Sciences, University of Copenhagen, Copenhagen, Denmark; 2Genetics and Bioinformatics, Department of Animal and Veterinary Basic Sciences, Faculty of Life Sciences, University of Copenhagen, Copenhagen, Denmark and 3Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio, TX, USA Aggregated distributions of macroparasites within their host 0.32–0.73 of the phenotypic variation for T. suis could be populations are characteristic of most natural and experi- attributed to genetic factors. For A. suum, heritabilities of mental infections. We designed this study to measure the 0.29–0.31 were estimated for log (FEC þ 1) at weeks 7–14 amount of variation that is attributable to host genetic factors p.i., whereas the heritability of log worm counts was 0.45. in a pig–helminth system. In total, 195 piglets were produced Strong positive genetic correlations (0.75–0.89) between after artificial insemination of 19 sows (Danish Landrace– T. suis and A. suum FECs suggest that resistance to both Yorkshire crossbreds) with semen selected from 13 indivi- infections involves regulation by overlapping genes. Our data dual Duroc boars (1 or 2 sows per boar; mean litter size: demonstrate that there is a strong genetic component in 10.3; 5–14 piglets per litter). -

Ascaris Lumbricoides and Strongyloides Stercoralis Associated Diarrhoea in an Immuno-Compromised Patient
IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS) e-ISSN:2278-3008, p-ISSN:2319-7676. Volume 11, Issue 5 Ver. IV (Sep. - Oct.2016), PP 29-32 www.iosrjournals.org Ascaris lumbricoides and Strongyloides stercoralis associated diarrhoea in an immuno-compromised patient Haodijam Ranjana1, Laitonjam Anand 2 and R.K.Gambhir Singh3 1 PhD student, Parasitology Section, Department of Life Sciences, Manipur University, Canchipur – 795 003, Imphal, Manipur (India) 2 Research Officer, Molecular Diagnostic Laboratory, Department of Microbiology, Regional Institute of Medical Sciences, Lamphelpat – 795 004, Imphal, Manipur (India) 3 Professor, Parasitology Section, Department of Life Sciences, Manipur University, Canchipur – 795 003, Imphal, Manipur (India) Abstract: As a part of ongoing research work on the prevalence and epidemiology of enteric parasites associated with HIV/AIDS patients, field visits were made in the Churachandpur district of Manipur during the period of February to May 2016, with a view to assess the occurrence/prevalence of opportunistic parasites in these immuno-compromised group of patients. During this field visit, a 40 year old HIV seropositive female, who worked as an outreach worker in one of the drug de-addiction centres, complained of experiencing diarrhoea since two and half months back. She also gave a history of loose motion/intermittent diarrhoea, on and off for the past 1-2 years. On laboratory investigation, using the standard parasitological techniques, she was diagnosed as suffering from Ascaris lumbricoides and Strongyloides stercoralis infection. Single infection either with Ascaris lumbricoides or Strongyloides stercoralis is of common occurrence, however concurrent infection with these two parasites is of infrequent occurrence. -

Preclinical Evaluation of Protein Disulfide Isomerase Inhibitors for the Treatment of Glioblastoma by Andrea Shergalis
Preclinical Evaluation of Protein Disulfide Isomerase Inhibitors for the Treatment of Glioblastoma By Andrea Shergalis A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Medicinal Chemistry) in the University of Michigan 2020 Doctoral Committee: Professor Nouri Neamati, Chair Professor George A. Garcia Professor Peter J. H. Scott Professor Shaomeng Wang Andrea G. Shergalis [email protected] ORCID 0000-0002-1155-1583 © Andrea Shergalis 2020 All Rights Reserved ACKNOWLEDGEMENTS So many people have been involved in bringing this project to life and making this dissertation possible. First, I want to thank my advisor, Prof. Nouri Neamati, for his guidance, encouragement, and patience. Prof. Neamati instilled an enthusiasm in me for science and drug discovery, while allowing me the space to independently explore complex biochemical problems, and I am grateful for his kind and patient mentorship. I also thank my committee members, Profs. George Garcia, Peter Scott, and Shaomeng Wang, for their patience, guidance, and support throughout my graduate career. I am thankful to them for taking time to meet with me and have thoughtful conversations about medicinal chemistry and science in general. From the Neamati lab, I would like to thank so many. First and foremost, I have to thank Shuzo Tamara for being an incredible, kind, and patient teacher and mentor. Shuzo is one of the hardest workers I know. In addition to a strong work ethic, he taught me pretty much everything I know and laid the foundation for the article published as Chapter 3 of this dissertation. The work published in this dissertation really began with the initial identification of PDI as a target by Shili Xu, and I am grateful for his advice and guidance (from afar!). -
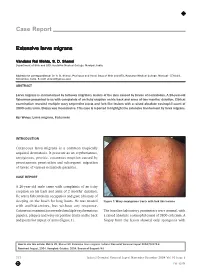
Extensive Larva Migrans
Case Report Extensive larva migrans Vandana Rai Mehta, S. D. Shenoi Department of Skin and STD, Kasturba Medical College, Manipal, India. Address for correspondence: Dr. S. D. Shenoi, Professor and Head, Dept of Skin and STD, Kasturba Medical College, Manipal - 576104, Karnataka, India. E-mail: [email protected] ABSTRACT Larva migrans is characterized by tortuous migratory lesions of the skin caused by larvae of nematodes. A 26-year-old fisherman presented to us with complaints of an itchy eruption on his back and arms of two months’ duration. Clinical examination revealed multiple wavy serpentine tracts and fork like lesions with a raised absolute eosinophil count of 3800 cells/cmm. Biopsy was inconclusive. This case is reported to highlight the extensive involvement by larva migrans. KEY WORDS: Larva migrans, Fisherman INTRODUCTION Cutaneous larva migrans is a common tropically acquired dermatosis. It presents as an erythematous, serpiginous, pruritic, cutaneous eruption caused by percutaneous penetration and subsequent migration of larvae of various nematode parasites. CASE REPORT A 26-year-old male came with complaints of an itchy eruption on his back and arms of 2 months’ duration. He was a fisherman by occupation and gave a history of sleeping on the beach for long hours. He was treated Figure 1: Wavy serpiginous tracts with fork like lesions with antihistamines, but without any response. Cutaneous examination revealed multiple erythematous The baseline laboratory parameters were normal, with papules, plaques and wavy serpentine tracts on the back a raised absolute eosinophil count of 3800 cell/cmm. A and posterior aspect of arms (Figure 1). biopsy from the lesion showed only spongiosis with How to cite this article: Mehta VR, Shenoi SD. -

February 15, 2012 Chapter 34 Notes: Flatworms, Roundworms and Rotifers
February 15, 2012 Chapter 34 Notes: Flatworms, Roundworms and Rotifers Section 1 Platyhelminthes Section 2 Nematoda and Rotifera 34-1 Objectives Summarize the distinguishing characteristics of flatworms. Describe the anatomy of a planarian. Compare free-living and parasitic flatworms. Diagram the life cycle of a fluke. Describe the life cycle of a tapeworm. Structure and Function of Flatworms · The phylum Platyhelminthes includes organisms called flatworms. · They are more complex than sponges but are the simplest animals with bilateral symmetry. · Their bodies develop from three germ layers: · ectoderm · mesoderm · endoderm · They are acoelomates with dorsoventrally flattened bodies. · They exhibit cephalization. · The classification of Platyhelminthes has undergone many recent changes. Characteristics of Flatworms February 15, 2012 Class Turbellaria · The majority of species in the class Turbellaria live in the ocean. · The most familiar turbellarians are the freshwater planarians of the genus Dugesia. · Planarians have a spade-shaped anterior end and a tapered posterior end. Class Turbellaria Continued Digestion and Excretion in Planarians · Planarians feed on decaying plant or animal matter and smaller organisms. · Food is ingested through the pharynx. · Planarians eliminate excess water through a network of excretory tubules. · Each tubule is connected to several flame cells. · The water is transported through the tubules and excreted from pores on the body surface. Class Turbellaria Continued Neural Control in Planarians · The planarian nervous system is more complex than the nerve net of cnidarians. · The cerebral ganglia serve as a simple brain. · A planarian’s nervous system gives it the ability to learn. · Planarians sense light with eyespots. · Other sensory cells respond to touch, water currents, and chemicals in the environment.