Epistatic Effects of Potassium Channel Variation on Cardiac Repolarization and Atrial Fibrillation Risk
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Potassium Channels in Epilepsy
Downloaded from http://perspectivesinmedicine.cshlp.org/ on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Potassium Channels in Epilepsy Ru¨diger Ko¨hling and Jakob Wolfart Oscar Langendorff Institute of Physiology, University of Rostock, Rostock 18057, Germany Correspondence: [email protected] This review attempts to give a concise and up-to-date overview on the role of potassium channels in epilepsies. Their role can be defined from a genetic perspective, focusing on variants and de novo mutations identified in genetic studies or animal models with targeted, specific mutations in genes coding for a member of the large potassium channel family. In these genetic studies, a demonstrated functional link to hyperexcitability often remains elusive. However, their role can also be defined from a functional perspective, based on dy- namic, aggravating, or adaptive transcriptional and posttranslational alterations. In these cases, it often remains elusive whether the alteration is causal or merely incidental. With 80 potassium channel types, of which 10% are known to be associated with epilepsies (in humans) or a seizure phenotype (in animals), if genetically mutated, a comprehensive review is a challenging endeavor. This goal may seem all the more ambitious once the data on posttranslational alterations, found both in human tissue from epilepsy patients and in chronic or acute animal models, are included. We therefore summarize the literature, and expand only on key findings, particularly regarding functional alterations found in patient brain tissue and chronic animal models. INTRODUCTION TO POTASSIUM evolutionary appearance of voltage-gated so- CHANNELS dium (Nav)andcalcium (Cav)channels, Kchan- nels are further diversified in relation to their otassium (K) channels are related to epilepsy newer function, namely, keeping neuronal exci- Psyndromes on many different levels, ranging tation within limits (Anderson and Greenberg from direct control of neuronal excitability and 2001; Hille 2001). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supporting Figure 2
Normalized value Functional class Genbank Name Naive SC 1h SC 6h SC 24h WM 1h WM 6h WM 24h AB016161 GABA-B receptor 1d AF109405 GABA-B receptor 2a M35077 Dopamine-1A receptor S46131 Dopamine-1A receptor M84009 Dopamine receptor D4 U13368 Adrenergic receptor, alpha 1a G Protein-coupled M60654 Adrenergic receptor, alpha 1d receptors and their M64236 Tachykinin 1 receptor effectors AI229237 Opioid receptor-like Y11433 Pyrimidinergic receptor P2Y4 M64299 Adenosine A1 receptor E00001 Pro-insulin M29014 Insulin receptor precursor U35315 Serotonin receptor 2C S62043 Serotonin receptor 6 AF000368 Scn9a sodium channel, type IX, alpha polypeptide M27158 Kcna5 K+ voltage-gated channel, shaker-related subfamily, member 5 X17621 Kcna6 potassium voltage-gated channel, shaker-related, subfamily, member 6 X16476 Kcnb1 potassium voltage gated channel, Shab-related subfamily, member 1 M77482 Kcnb2 potassium voltage gated channel, Shab-related subfamily, member 2 S64320 Kcnd2 potassium voltage gated channel, Shal-related family, member 2 Ion channels X87635 Kcnj4 potassium inwardly-rectifying channel, subfamily J, member 4 D86039 Kcnj11 potassium inwardly-rectifying channel, subfamily J, member 11 X83581 Kcnj16 potassium inwardly-rectifying channel, subfamily J, member 16 AF073891 Kcnh5 potassium voltage-gated channel, subfamily H (eag-related), member 5 U69882 Kcnn2 potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 Z36944 Chloride channel 4-2 Z56277 Chloride channel 5 L08493 GABA-A receptor alpha-4 subunit X51992 -

The Methamphetamine-Induced RNA Targetome of Hnrnp H in Hnrnph1 Mutants Showing Reduced Dopamine Release and Behavior
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.06.451358; this version posted July 7, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The methamphetamine-induced RNA targetome of hnRNP H in Hnrnph1 mutants showing reduced dopamine release and behavior Qiu T. Ruan1,2,3, Michael A. Rieger4, William B. Lynch2,5, Jiayi W. Cox6, Jacob A. Beierle1,2,3, Emily J. Yao2, Amarpreet Kandola2, Melanie M. Chen2, Julia C. Kelliher2, Richard K. Babbs2, Peter E. A. Ash7, Benjamin Wolozin7, Karen K. Szumlinski8, W. Evan Johnson9, Joseph D. Dougherty4, and Camron D. Bryant1,2,3* 1. Biomolecular Pharmacology Training Program, Department of Pharmacology and Experimental Therapeutics, Boston University School of Medicine 2. Laboratory of Addiction Genetics, Department of Pharmacology and Experimental Therapeutics and Psychiatry, Boston University School of Medicine 3. Transformative Training Program in Addiction Science, Boston University School of Medicine 4. Department of Genetics, Department of Psychiatry, Washington University School of Medicine 5. Graduate Program for Neuroscience, Boston University 6. Programs in Biomedical Sciences, Boston University School of Medicine 7. Department of Pharmacology and Experimental Therapeutics and Neurology, Boston University School of Medicine 8. Department of Psychological and Brain Sciences, University of California, Santa Barbara 9. Department of Medicine, Computational Biomedicine, Boston University School of Medicine *Corresponding Author Camron D. Bryant, Ph.D. Department of Pharmacology and Experimental Therapeutics and Department of Psychiatry 72 E. -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. -

Dimethylation of Histone 3 Lysine 9 Is Sensitive to the Epileptic Activity
1368 MOLECULAR MEDICINE REPORTS 17: 1368-1374, 2018 Dimethylation of Histone 3 Lysine 9 is sensitive to the epileptic activity, and affects the transcriptional regulation of the potassium channel Kcnj10 gene in epileptic rats SHAO-PING ZHANG1,2*, MAN ZHANG1*, HONG TAO1, YAN LUO1, TAO HE3, CHUN-HUI WANG3, XIAO-CHENG LI3, LING CHEN1,3, LIN-NA ZHANG1, TAO SUN2 and QI-KUAN HU1-3 1Department of Physiology; 2Ningxia Key Laboratory of Cerebrocranial Diseases, Incubation Base of National Key Laboratory, Ningxia Medical University; 3General Hospital of Ningxia Medical University, Yinchuan, Ningxia 750004, P.R. China Received February 18, 2017; Accepted September 13, 2017 DOI: 10.3892/mmr.2017.7942 Abstract. Potassium channels can be affected by epileptic G9a by 2-(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-di- seizures and serve a crucial role in the pathophysiology of methoxy-N-(1-(phenyl-methyl)-4-piperidinyl)-4-quinazolinamine epilepsy. Dimethylation of histone 3 lysine 9 (H3K9me2) and tri-hydrochloride hydrate (bix01294) resulted in upregulation its enzyme euchromatic histone-lysine N-methyltransferase 2 of the expression of Kir4.1 proteins. The present study demon- (G9a) are the major epigenetic modulators and are associated strated that H3K9me2 and G9a are sensitive to epileptic seizure with gene silencing. Insight into whether H3K9me2 and G9a activity during the acute phase of epilepsy and can affect the can respond to epileptic seizures and regulate expression of transcriptional regulation of the Kcnj10 channel. genes encoding potassium channels is the main purpose of the present study. A total of 16 subtypes of potassium channel Introduction genes in pilocarpine-modelled epileptic rats were screened by reverse transcription-quantitative polymerase chain reac- Epilepsies are disorders of neuronal excitability, characterized tion, and it was determined that the expression ATP-sensitive by spontaneous and recurrent seizures. -

Pflugers Final
CORE Metadata, citation and similar papers at core.ac.uk Provided by Serveur académique lausannois A comprehensive analysis of gene expression profiles in distal parts of the mouse renal tubule. Sylvain Pradervand2, Annie Mercier Zuber1, Gabriel Centeno1, Olivier Bonny1,3,4 and Dmitri Firsov1,4 1 - Department of Pharmacology and Toxicology, University of Lausanne, 1005 Lausanne, Switzerland 2 - DNA Array Facility, University of Lausanne, 1015 Lausanne, Switzerland 3 - Service of Nephrology, Lausanne University Hospital, 1005 Lausanne, Switzerland 4 – these two authors have equally contributed to the study to whom correspondence should be addressed: Dmitri FIRSOV Department of Pharmacology and Toxicology, University of Lausanne, 27 rue du Bugnon, 1005 Lausanne, Switzerland Phone: ++ 41-216925406 Fax: ++ 41-216925355 e-mail: [email protected] and Olivier BONNY Department of Pharmacology and Toxicology, University of Lausanne, 27 rue du Bugnon, 1005 Lausanne, Switzerland Phone: ++ 41-216925417 Fax: ++ 41-216925355 e-mail: [email protected] 1 Abstract The distal parts of the renal tubule play a critical role in maintaining homeostasis of extracellular fluids. In this review, we present an in-depth analysis of microarray-based gene expression profiles available for microdissected mouse distal nephron segments, i.e., the distal convoluted tubule (DCT) and the connecting tubule (CNT), and for the cortical portion of the collecting duct (CCD) (Zuber et al., 2009). Classification of expressed transcripts in 14 major functional gene categories demonstrated that all principal proteins involved in maintaining of salt and water balance are represented by highly abundant transcripts. However, a significant number of transcripts belonging, for instance, to categories of G protein-coupled receptors (GPCR) or serine-threonine kinases exhibit high expression levels but remain unassigned to a specific renal function. -

LGI1–ADAM22–MAGUK Configures Transsynaptic Nanoalignment for Synaptic Transmission and Epilepsy Prevention
LGI1–ADAM22–MAGUK configures transsynaptic nanoalignment for synaptic transmission and epilepsy prevention Yuko Fukataa,b,1, Xiumin Chenc,d,1, Satomi Chikenb,e, Yoko Hiranoa,f, Atsushi Yamagatag, Hiroki Inahashia, Makoto Sanboh, Hiromi Sanob,e, Teppei Gotoh, Masumi Hirabayashib,h, Hans-Christian Kornaui,j, Harald Prüssi,k, Atsushi Nambub,e, Shuya Fukail, Roger A. Nicollc,d,2, and Masaki Fukataa,b,2 aDivision of Membrane Physiology, Department of Molecular and Cellular Physiology, National Institute for Physiological Sciences, National Institutes of Natural Sciences, Aichi 444-8787, Japan; bDepartment of Physiological Sciences, School of Life Science, SOKENDAI (The Graduate University for Advanced Studies), Aichi 444-8585, Japan; cDepartment of Cellular and Molecular Pharmacology, University of California, San Francisco, CA 94158; dDepartment of Physiology, University of California, San Francisco, CA 94158; eDivision of System Neurophysiology, Department of System Neuroscience, National Institute for Physiological Sciences, National Institutes of Natural Sciences, Aichi 444-8585, Japan; fDepartment of Pediatrics, Graduate School of Medicine, The University of Tokyo, Tokyo 113-8655, Japan; gLaboratory for Protein Functional and Structural Biology, RIKEN Center for Biosystems Dynamics Research, Kanagawa 230-0045, Japan; hCenter for Genetic Analysis of Behavior, National Institute for Physiological Sciences, National Institutes of Natural Sciences, Okazaki 444-8787, Japan; iGerman Center for Neurodegenerative Diseases (DZNE), 10117 Berlin, Germany; jNeuroscience Research Center, Cluster NeuroCure, Charité-Universitätsmedizin Berlin, 10117 Berlin, Germany; kDepartment of Neurology and Experimental Neurology, Charité-Universitätsmedizin Berlin, 10117 Berlin, Germany; and lDepartment of Chemistry, Graduate School of Science, Kyoto University, 606-8502 Kyoto, Japan Contributed by Roger A. Nicoll, December 1, 2020 (sent for review October 29, 2020; reviewed by David S. -
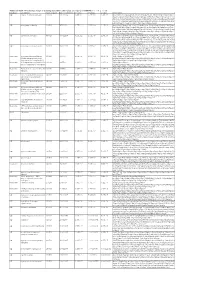
Additional Tables.Xlsx
Additional Table 6 Enriched pathways of downregulated DEGs after spinal cord injury in CRMP2KI (P < 0.05, q < 0.05) Database Description Ratio of DEGs Ratio of universe P -value P adjust q value Gene name GO neuron to neuron synapse 52/523 379/11234 1.49E-12 7.29E-10 5.98E-10 Drp2/Grik5/Tnr/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/ Neurl1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/ Adam10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/P alm/Slc30a3/Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/R pl8/Nsf/Rps19/Camk2n1/Syt11/Syt1/Rps14/Nrgn/Rpl38 GO postsynaptic density 49/523 357/11234 6.77E-12 1.51E-09 1.24E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/Neur l1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/Ada m10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/Palm/ Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/Rpl8/Nsf/Rps1 9/Camk2n1/Syt11/Rps14/Nrgn/Rpl38 GO asymmetric synapse 49/523 360/11234 9.23E-12 1.51E-09 1.24E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/Neur l1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/Ada m10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/Palm/ Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/Rpl8/Nsf/Rps1 9/Camk2n1/Syt11/Rps14/Nrgn/Rpl38 GO postsynaptic specialization 50/523 382/11234 2.41E-11 2.95E-09 2.42E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Dlgap3/Slitrk1/Arrb1/Add3/Cdk l5/Neurl1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4 -

Altered Enhancer Transcription Underlies Huntington's Disease Striatal Transcriptional Signature
www.nature.com/scientificreports OPEN Altered enhancer transcription underlies Huntington’s disease striatal transcriptional signature Received: 02 August 2016 Stéphanie Le Gras1,*, Céline Keime1,*, Anne Anthony2,3,*, Caroline Lotz2,3, Accepted: 16 January 2017 Lucie De Longprez4,5, Emmanuel Brouillet4,5, Jean-Christophe Cassel2,3, Published: 22 February 2017 Anne-Laurence Boutillier2,3 & Karine Merienne2,3 Epigenetic and transcriptional alterations are both implicated in Huntington’s disease (HD), a progressive neurodegenerative disease resulting in degeneration of striatal neurons in the brain. However, how impaired epigenetic regulation leads to transcriptional dysregulation in HD is unclear. Here, we investigated enhancer RNAs (eRNAs), a class of long non-coding RNAs transcribed from active enhancers. We found that eRNAs are expressed from many enhancers of mouse striatum and showed that a subset of those eRNAs are deregulated in HD vs control mouse striatum. Enhancer regions producing eRNAs decreased in HD mouse striatum were associated with genes involved in striatal neuron identity. Consistently, they were enriched in striatal super-enhancers. Moreover, decreased eRNA expression in HD mouse striatum correlated with down-regulation of associated genes. Additionally, a significant number of RNA Polymerase II (RNAPII) binding sites were lost within enhancers associated with decreased eRNAs in HD vs control mouse striatum. Together, this indicates that loss of RNAPII at HD mouse enhancers contributes to reduced transcription of eRNAs, resulting in down-regulation of target genes. Thus, our data support the view that eRNA dysregulation in HD striatum is a key mechanism leading to altered transcription of striatal neuron identity genes, through reduced recruitment of RNAPII at super-enhancers. -

Disease Associated Mutations in KIR Proteins Linked to Aberrant Inward Rectifier Channel Trafficking
biomolecules Review Disease Associated Mutations in KIR Proteins Linked to Aberrant Inward Rectifier Channel Trafficking 1, 2, 2 1 Eva-Maria Zangerl-Plessl y, Muge Qile y, Meye Bloothooft , Anna Stary-Weinzinger and Marcel A. G. van der Heyden 2,* 1 Department of Pharmacology and Toxicology, University of Vienna, 1090 Vienna, Austria; [email protected] (E.-M.Z.-P.); [email protected] (A.S.-W.) 2 Department of Medical Physiology, Division of Heart & Lungs, University Medical Center Utrecht, 3584 CM Utrecht, The Netherlands; [email protected] (M.Q.); [email protected] (M.B.) * Correspondence: [email protected]; Tel.: +31-887558901 These authors contributed equally to this work. y Received: 28 August 2019; Accepted: 23 October 2019; Published: 25 October 2019 Abstract: The ubiquitously expressed family of inward rectifier potassium (KIR) channels, encoded by KCNJ genes, is primarily involved in cell excitability and potassium homeostasis. Channel mutations associate with a variety of severe human diseases and syndromes, affecting many organ systems including the central and peripheral neural system, heart, kidney, pancreas, and skeletal muscle. A number of mutations associate with altered ion channel expression at the plasma membrane, which might result from defective channel trafficking. Trafficking involves cellular processes that transport ion channels to and from their place of function. By alignment of all KIR channels, and depicting the trafficking associated mutations, three mutational hotspots were identified. One localized in the transmembrane-domain 1 and immediately adjacent sequences, one was found in the G-loop and Golgi-export domain, and the third one was detected at the immunoglobulin-like domain. -

Jcvtr-30194.Pdf
J Cardiovasc Thorac Res, 2021, 13(x), x-x doi: 10.34172/jcvtr.2021.32 TUOMS http://jcvtr.tbzmed.ac.ir P R E S S Original Article Gender-specific characteristics of hypertrophic response in cardiomyocytes derived from human embryonic stem cells Shiva Ahmadvand1 ID , Ali Osia2, Anna Meyfour3, Sara Pahlavan1* ID 1Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran 2Cobel Darou, Tehran, Iran 3Basic and Molecular Epidemiology of Gastrointestinal Disorders Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran Article info Abstract Article History: Introduction: Gender-specific phenotypes of the heart were reported with respect to both physiology Received: 2 February 2021 and pathology. While most differences were associated with the sex hormones, differential expression Accepted: 30 April 2021 of genes received special attention, particularly X-Y chromosomes’ genes. epublished: 16 May 2021 Methods: Here, we compared cardiogenesis by gene expression analysis of lineage specific markers and X-Y chromosomes’ genes, during in vitro differentiation of XY and XX human embryonic stem cells Keywords: (hESC), in a hormone-free setup. Embryonic Stem Cells Results: Downregulation of pluripotency marker (NANOG) and upregulation of cardiac mesoderm and Cardiomyocyte Differentiation progenitor markers (GATA4, TBX5, NKX2.5, ISL1) was remained temporally similar in differentiating Sexual Dimorphism XY and XX hESCs. Isoproterenol treatment of XY and XX hESC-derived cardiomyocytes (hESC- Hypertrophy CM) induced hypertrophy in a sex-specific manner, with female cardiomyocytes showing response at higher isoproterenol concentration and a later time point of differentiation.