Pseudo-Repeats in Doublecortin Make Distinct Mechanistic Contributions to Microtubule Regulation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genotype-Phenotype Correlations in RP1-Associated Retinal Dystrophies: a Multi-Center Cohort Study in JAPAN
Journal of Clinical Medicine Article Genotype-Phenotype Correlations in RP1-Associated Retinal Dystrophies: A Multi-Center Cohort Study in JAPAN Kei Mizobuchi 1,* , Takaaki Hayashi 1,2 , Noriko Oishi 3, Daiki Kubota 3, Shuhei Kameya 3, Koichiro Higasa 4, Takuma Futami 5, Hiroyuki Kondo 5 , Katsuhiro Hosono 6, Kentaro Kurata 6, Yoshihiro Hotta 6 , Kazutoshi Yoshitake 7, Takeshi Iwata 7 , Tomokazu Matsuura 8 and Tadashi Nakano 1 1 Department of Ophthalmology, The Jikei University School of Medicine, 3-19-18, Nishi-shimbashi, Minato-ku, Tokyo 105-8471, Japan; [email protected] (T.H.); [email protected] (T.N.) 2 Department of Ophthalmology, Katsushika Medical Center, The Jikei University School of Medicine, 6-41-2 Aoto, Katsushika-ku, Tokyo 125-8506, Japan 3 Department of Ophthalmology, Nippon Medical School Chiba Hokusoh Hospital, 1715 Kamagari, Inzai, Chiba 270-1694, Japan; [email protected] (N.O.); [email protected] (D.K.); [email protected] (S.K.) 4 Department of Genome Analysis, Institute of Biomedical Science, Kansai Medical University, 2-5-1 Shinmachi, Hirakata, Osaka 573-1010, Japan; [email protected] 5 Department of Ophthalmology, University of Occupational and Environmental Health, 1-1, Iseigaoka, Yahatanishi-ku Kitakyushu-shi, Fu-kuoka 807-8555, Japan; [email protected] (T.F.); [email protected] (H.K.) 6 Department of Ophthalmology, Hamamatsu University School of Medicine, 1-20-1, Handayama, Higashi-ku, Shizuoka, Hamamatsu 431-3192, Japan; [email protected] (K.H.); [email protected] (K.K.); [email protected] (Y.H.) 7 Citation: Mizobuchi, K.; Hayashi, T.; National Institute of Sensory Organs, National Hospital Organization Tokyo Medical Center, Oishi, N.; Kubota, D.; Kameya, S.; 2-5-1 Higashigaoka, Meguro-ku, Tokyo 152-8902, Japan; [email protected] (K.Y.); [email protected] (T.I.) Higasa, K.; Futami, T.; Kondo, H.; 8 Department of Laboratory Medicine, The Jikei University School of Medicine, 3-19-18, Nishi-shimbashi, Hosono, K.; Kurata, K.; et al. -

Mechanisms and Disturbances of Neuronal Migration the Developing Nervous System: a Series of Review Articles
0031-3998/00/4806-0725 PEDIATRIC RESEARCH Vol. 48, No. 6, 2000 Copyright © 2000 International Pediatric Research Foundation, Inc. Printed in U.S.A. The Developing Nervous System: A Series of Review Articles The following article is the first in an important series of review articles which discuss the developmental biology of the nervous system and its relation to diseases and disorders that are found in new born infants and children. This review, by Pierre Gressens, describes neuronal migration in the developing brain and abnormalities in humans resulting from aberrations in neuronal migration. Alvin Zipursky Editor-in-Chief Mechanisms and Disturbances of Neuronal Migration PIERRE GRESSENS Service de Neurologie Pédiatrique and INSERM E 9935, Hôpital Robert-Debré, F-75019 Paris, France ABSTRACT Neuronal migration appears as a complex ontogenic step of migration of neurons destined to the neocortex. New insights occurring early during embryonic and fetal development. Control gained from the analysis of animal models as well as from the of neuronal migration involves different cell populations includ- study of human diseases will be included. (Pediatr Res 48: ing Cajal-Retzius neurons, subplate neurons, neuronal precursors 725–730, 2000) or radial glia. The integrity of multiple molecular mechanisms, such as cell cycle control, cell-cell adhesion, interaction with extracellular matrix protein, neurotransmitter release, growth Abbreviations: factor availability, platelet-activating factor degradation or trans- BDNF, brain-derived neurotrophic factor duction pathways seems to be critical for normal neuronal mi- EGF, epidermal growth factor gration. The complexity and the multiplicity of these mecha- NMDA, N-methyl-D-aspartate nisms probably explain the clinical, radiologic and genetic NT, neurotrophin heterogeneity of human disorders of neuronal migration. -

Doublecortin (Dcx) Family Proteins Regulate Filamentous Actin Structure in Developing Neurons
The Journal of Neuroscience, January 9, 2013 • 33(2):709–721 • 709 Cellular/Molecular Doublecortin (Dcx) Family Proteins Regulate Filamentous Actin Structure in Developing Neurons Xiaoqin Fu,1 Kristy J. Brown,2 Chan Choo Yap,3 Bettina Winckler,3 Jyoti K. Jaiswal,2 and Judy S. Liu1 1Center for Neuroscience Research and 2Center for Genetic Medicine Research, Children’s National Medical Center, Washington, DC 20010, and 3Department of Neuroscience, University of Virginia Medical School, Charlottesville, Virginia 22908 Doublecortin (Dcx) is the causative gene for X-linked lissencephaly, which encodes a microtubule-binding protein. Axon tracts are abnormal in both affected individuals and in animal models. To determine the reason for the axon tract defect, we performed a semi- quantitative proteomic analysis of the corpus callosum in mice mutant for Dcx. In axons from mice mutant for Dcx, widespread differ- ences are found in actin-associated proteins as compared with wild-type axons. Decreases in actin-binding proteins ␣-actinin-1 and ␣-actinin-4 and actin-related protein 2/3 complex subunit 3 (Arp3), are correlated with dysregulation in the distribution of filamentous actin (F-actin) in the mutant neurons with increased F-actin around the cell body and decreased F-actin in the neurites and growth cones. The actin distribution defect can be rescued by full-length Dcx and further enhanced by Dcx S297A, the unphosphorylatable mutant, but not with the truncation mutant of Dcx missing the C-terminal S/P-rich domain. Thus, the C-terminal region of Dcx dynamically regulates formation of F-actin features in developing neurons, likely through interaction with spinophilin, but not through ␣-actinin-4 or Arp3. -

The Retinitis Pigmentosa 1 Protein Is a Photoreceptor Microtubule-Associated Protein
The Journal of Neuroscience, July 21, 2004 • 24(29):6427–6436 • 6427 Neurobiology of Disease The Retinitis Pigmentosa 1 Protein Is a Photoreceptor Microtubule-Associated Protein Qin Liu,1 Jian Zuo,2 and Eric A. Pierce1 1F. M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104, and 2Department of Developmental Neurobiology, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105 The outer segments of rod and cone photoreceptor cells are highly specialized sensory cilia made up of hundreds of membrane discs stacked into an orderly array along the photoreceptor axoneme. It is not known how the alignment of the outer segment discs is controlled, although it has been suggested that the axoneme may play a role in this process. Mutations in the retinitis pigmentosa 1 (RP1) gene are a common cause of retinitis pigmentosa (RP). Disruption of the Rp1 gene in mice causes misorientation of outer segment discs, suggesting a role for RP1 in outer segment organization. Here, we show that the RP1 protein is part of the photoreceptor axoneme. Amino acids 28–228 of RP1, which share limited homology with the microtubule-binding domains of the neuronal microtubule-associated protein (MAP) doublecortin, mediate the interaction between RP1 and microtubules, indicating that the putative doublecortin (DCX) domains in RP1 are functional. The N-terminal portion of RP1 stimulates the formation of microtubules in vitro and stabilizes cytoplas- mic microtubules in heterologous cells. Evaluation of photoreceptor axonemes from mice with targeted disruptions of the Rp1 gene shows that Rp1 proteins that contain the DCX domains also help control axoneme length and stability in vivo. -

Doublecortin Is Required in Mice for Lamination of the Hippocampus but Not the Neocortex
The Journal of Neuroscience, September 1, 2002, 22(17):7548–7557 Doublecortin Is Required in Mice for Lamination of the Hippocampus But Not the Neocortex Joseph C. Corbo,1,2 Thomas A. Deuel,1 Jeffrey M. Long,3 Patricia LaPorte,3 Elena Tsai,1 Anthony Wynshaw-Boris,3 and Christopher A. Walsh1 1Department of Neurology, Beth Israel Deaconess Medical Center, and Programs in Neuroscience and Biological and Biomedical Sciences, Harvard Medical School, Boston, Massachusetts 02115, 2Department of Pathology, Brigham and Women’s Hospital, Boston, Massachusetts 02115, and 3Departments of Pediatrics and Medicine, University of California, San Diego School of Medicine, San Diego, California 92093 Doublecortin (DCX) is a microtubule-associated protein that is cal lamination that is largely indistinguishable from wild type required for normal neocortical and hippocampal development and show normal patterns of neocortical neurogenesis and in humans. Mutations in the X-linked human DCX gene cause neuronal migration. In contrast, the hippocampus of both het- gross neocortical disorganization (lissencephaly or “smooth erozygous females and hemizygous males shows disrupted brain”) in hemizygous males, whereas heterozygous females lamination that is most severe in the CA3 region. Behavioral show a mosaic phenotype with a normal cortex as well as a tests show defects in context and cued conditioned fear tests, second band of misplaced (heterotopic) neurons beneath the suggesting that deficits in hippocampal learning accompany cortex (“double cortex syndrome”). We created a mouse carry- the abnormal cytoarchitecture. ing a targeted mutation in the Dcx gene. Hemizygous male Dcx mice show severe postnatal lethality; the few that survive to Key words: doublecortin; knock-out; mouse; cerebral cortex; adulthood are variably fertile. -

Apoer2/VLDL Receptor and Dab1 in the Rostral Migratory Stream Function in Postnatal Neuronal Migration Independently of Reelin
ApoER2/VLDL receptor and Dab1 in the rostral migratory stream function in postnatal neuronal migration independently of Reelin Nuno Andrade*, Vukoslav Komnenovic†, Sophia M. Blake*, Yves Jossin‡, Brian Howell§, Andre Goffinet‡, Wolfgang J. Schneider*, and Johannes Nimpf*¶ *Max F. Perutz Laboratories, University Departments at the Vienna Biocenter, Department of Medical Biochemistry, Medical University of Vienna, A-1030 Vienna, Austria; †Institute of Molecular Biotechnology, Austrian Academy of Sciences, 1030 Vienna, Austria; ‡Developmental Neurobiology Unit, University of Leuven Medical School, 3000 Leuven, Belgium; and §Neurogenetics Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892 Edited by Thomas C. Su¨dhof, The University of Texas Southwestern Medical Center, Dallas, TX, and approved March 30, 2007 (received for review December 21, 2006) Postnatal migration of interneuron precursors from the subventricu- In the cerebrum, Reelin is crucial for correct positioning of lar zone to the olfactory bulb occurs in chains that form the substrate radially migrating neuroblasts via its binding to ApoER2 and for the rostral migratory stream. Reelin is suggested to induce de- very-low-density lipoprotein receptor (VLDLR) (18, 19), which tachment of neuroblasts from the chains when they arrive at the triggers tyrosine phosphorylation of the adaptor Dab1 by re- olfactory bulb. Here we show that ApoER2 and possibly very-low- ceptor clustering (20). Binding of Reelin to the receptors and density lipoprotein receptor (VLDLR) and their intracellular adapter subsequent phosphorylation of Dab1 are consecutive steps of a protein Dab1 are involved in chain formation most likely independent linear pathway, because disruption of any of the corresponding of Reelin. -

Widely Used Genetic Technique May Lead to Spurious Results
Spectrum | Autism Research News https://www.spectrumnews.org NEWS Widely used genetic technique may lead to spurious results BY JESSICA WRIGHT 7 AUGUST 2014 Target practice: An RNA fragment designed to block the doublecortin gene misdirects neurons (right two panels) from their correct destinations (left) even in cells that lack the gene (far right). A popular method that neuroscientists use to simulate the effects of a mutation may alter the brain and produce misleading results, cautions a study published 18 June in Neuron1. The technique involves using a fragment of RNA, called a short hairpin RNA (shRNA), to interfere with protein production. It simulates the effects of a harmful mutation without having to change the genetic code, and is widely used because of its relative simplicity. For years, studies have hinted that the method may have ‘off-target’ effects — meaning that a given shRNA may block more than just its intended target. For example, a 2003 study found that an shRNA designed for the doublecortin gene prevents neurons from migrating to their correct locations in the mouse brain2. Deleting the gene in mice does not show this effect, suggesting that it is an unintended consequence3. In 2006, another study showed that a supposedly benign shRNA can weaken the connections between neurons4. “We had this controversy that existed for many years,” says Joseph Gleeson, professor of neurosciences at the University of California, San Diego, and lead researcher on the new study. “The neurobiology field continued to use shRNAs because they’re so easy to use, but there was this dark cloud hanging over the field that it might not be completely reliable.” 1 / 3 Spectrum | Autism Research News https://www.spectrumnews.org The new study confirms those fears. -

Dissertation
DISSERTATION Titel der Dissertation Characterization of Thrombospondin-1 as novel ligand for the Reelin pathway receptors ApoER2 and VLDLR in the postnatal mouse brain angestrebter akademischer Grad Doktor/in der Naturwissenschaften (Dr. rer.nat.) Verfasserin / Verfasser: Sophia Maria Blake Matrikel-Nummer: 0021434 Dissertationsgebiet (lt. Stu- A 091490 Molekulare Biologie dienblatt): Betreuerin / Betreuer: Univ.-Prof. Dr. Johannes Nimpf Wien, am 06. Dezember 2008 Formular Nr.: A.04 Contents 1 Contents 1 Abstract .................................................................................... 3 2 Zusammenfassung .................................................................. 5 3 Introduction .............................................................................. 7 3.1 Neocortical development ..................................................................7 3.1.1 The Neocortex............................................................................. 7 3.1.2 Neuronal migration in the developing neocortex .................... 8 3.1.3 Cortical layering........................................................................ 10 3.1.4 Mouse strains with defects in cortical architecture............... 11 3.1.4.1 reeler................................................................................... 11 3.1.4.2 dab1-/- (scrambler, yotari) .................................................. 11 3.1.4.3 vldlr-/-/apoER2-/- .................................................................. 12 3.2 The Reelin signaling cascade........................................................ -

In Vivo Conversion of Astrocytes to Neurons in the Injured Adult Spinal Cord
ARTICLE Received 2 Jan 2014 | Accepted 29 Jan 2014 | Published 25 Feb 2014 DOI: 10.1038/ncomms4338 In vivo conversion of astrocytes to neurons in the injured adult spinal cord Zhida Su1,2, Wenze Niu1, Meng-Lu Liu1, Yuhua Zou1 & Chun-Li Zhang1 Spinal cord injury (SCI) leads to irreversible neuronal loss and glial scar formation, which ultimately result in persistent neurological dysfunction. Cellular regeneration could be an ideal approach to replenish the lost cells and repair the damage. However, the adult spinal cord has limited ability to produce new neurons. Here we show that resident astrocytes can be converted to doublecortin (DCX)-positive neuroblasts by a single transcription factor, SOX2, in the injured adult spinal cord. Importantly, these induced neuroblasts can mature into synapse-forming neurons in vivo. Neuronal maturation is further promoted by treatment with a histone deacetylase inhibitor, valproic acid (VPA). The results of this study indicate that in situ reprogramming of endogenous astrocytes to neurons might be a potential strategy for cellular regeneration after SCI. 1 Department of Molecular Biology, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd, Dallas, Texas 75390-9148, USA. 2 Institute of Neuroscience and Key Laboratory of Molecular Neurobiology of Ministry of Education, Neuroscience Research Center of Changzheng Hospital, Second Military Medical University, 800 Xiangyin Rd, Shanghai 200433, China. Correspondence and requests for materials should be addressed to C.-L.Z. (email: [email protected]). -

The X-Linked Intellectual Disability Gene Product and E3 Ubiquitin Ligase KLHL15 Degrades Doublecortin Proteins to Constrain Neuronal Dendritogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.02.324285; this version posted October 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. KLHL15 degrades doublecortin proteins The X-linked intellectual disability gene product and E3 ubiquitin ligase KLHL15 degrades doublecortin proteins to constrain neuronal dendritogenesis Jianing Song1,2, Ronald A. Merrill1,2, Andrew Y. Usachev1, and Stefan Strack1* From the 1Department of Neuroscience and Pharmacology and the Iowa Neuroscience Institute, University of Iowa, Iowa City, Iowa 52242 2 These authors contributed equally to the work. *To whom correspondence should be addressed: Stefan Strack: Dept. of Neuroscience and Pharmacology, The University of Iowa, Iowa City, IA 52242; [email protected]; Tel. (319) 384-4439; Fax. (319) 335-8930 Running Title: KLHL15 degrades doublecortin proteins Keywords: Kelch-like 15, KLHL15, E3 ubiquitin ligase, protein phosphatase 2A, signal transduction, doublecortin, doublecortin-like kinases, microtubule-associated protein, ubiquitination, proteasomal degradation, protein turnover, neurite outgrowth, dendritic complexity, pulse-chase, HaloTag ______________________________________________________________________________ ABSTRACT doublecortin (DCX), also an X-linked disease gene, and doublecortin-like kinases 1 and 2 Proper brain development and function (DCLK1/2) as bona fide KLHL15 interactors requires finely controlled mechanisms -

82310507.Pdf
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Neuron, Vol. 23, 247±256, June, 1999, Copyright 1999 by Cell Press Doublecortin Is a Developmentally Regulated, Microtubule-Associated Protein Expressed in Migrating and Differentiating Neurons ² Fiona Francis,*k Annette Koulakoff, in the deeper layers, while later born neurons cross Dominique Boucher,² Philippe Chafey,* through these layers to successively reach regions Bruce Schaar,³ Marie-Claude Vinet,* closer to the cerebral surface (reviewed by McConnell, GaeÈ lle Friocourt,* Nathalie McDonnell,* 1995; Caviness et al., 1997). In this way, a six-layered Orly Reiner,§ Axel Kahn,* Susan K. McConnell,³ neocortex develops. Migrating neurons mainly travel in Yoheved Berwald-Netter,² Philippe Denoulet,² association with radially extending glial fibres (Rakic, and Jamel Chelly* 1971; Luskin et al., 1988); however, tangential and chain *U129 de l'INSERM migration have also been observed in some brain re- Institut Cochin de Ge ne tique Mole culaire gions (Luskin, 1993; Tan and Breen, 1993; O'Rourke et 24, rue du Faubourg Saint Jacques al., 1995; Lois et al., 1996). Many questions still remain 75014 Paris concerning the corticogenesis process: the signals that ² Laboratoire de Biochimie Cellulaire determine the timing of cells leaving the proliferative CNRS UPR 9065 pool, which ultimately affects their laminar fate (Frantz ColleÁ ge de France and McConnell, 1996), the mechanism and direction of 11, Place M. Berthelot cell locomotion, and the recognition of the final desti- 75005 Paris nation. France Lissencephaly is a cortical malformation disorder as- ³ Department of Biological Sciences sociated with severe mental retardation and epilepsy Stanford University (Harding, 1996), which exists as an X-linked disease (X-LIS) Stanford, California 94305 caused by mutations in the doublecortin gene (des § Department of Molecular Genetics Portes et al., 1998a; Gleeson et al., 1998). -
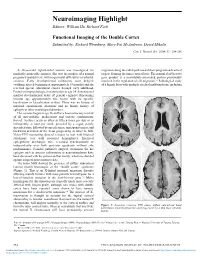
Functional Imaging of the Double Cortex Submitted By: Richard Wennberg, Mary-Pat Mcandrews, David Mikulis
Neuroimaging Highlight Editors: William Hu, Richard Farb Functional Imaging of the Double Cortex Submitted by: Richard Wennberg, Mary-Pat McAndrews, David Mikulis Can. J. Neurol. Sci. 2004; 31: 254-256 A 24-year-old right-handed woman was investigated for migration along the radial path toward their programmed cortical medically intractable seizures. She was the product of a normal targets, forming the inner cortical layer. The normal doublecortin pregnancy and delivery, with no perinatal difficulties or infantile gene product is a microtubule-associated protein presumably seizures. Early developmental milestones were delayed involved in the regulation of cell migration.5,6 Pathological study (walking, speech beginning at approximately 19 months) and she of a female fetus with multiple cerebral malformations, including received special educational classes through early adulthood. Formal neuropsychological examination at age 24 demonstrated marked developmental delay of general cognitive functioning (mental age approximately five years) with no specific localization or lateralization evident. There was no history of maternal spontaneous abortions and no family history of epilepsy or other neurological disorders. Her seizures began at age 16 and have been refractory to trials of all anti-epileptic medications and various combinations thereof. Seizures occur as often as fifteen times per day or as infrequently as once per week, preceded by a vague, poorly described aura, followed by speech arrest, unresponsiveness, and backward deviation of the head, progressing at times to falls. Video-EEG monitoring showed seizures to start with bilateral synchrony over both posterior hemispheres. Interictal epileptiform discharges were recorded synchronously or independently over both posterior quadrants without side predominance.