Early-Life Epilepsies and the Emerging Role of Genetic Testing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

IDF Patient & Family Handbook
Immune Deficiency Foundation Patient & Family Handbook for Primary Immunodeficiency Diseases This book contains general medical information which cannot be applied safely to any individual case. Medical knowledge and practice can change rapidly. Therefore, this book should not be used as a substitute for professional medical advice. FIFTH EDITION COPYRIGHT 1987, 1993, 2001, 2007, 2013 IMMUNE DEFICIENCY FOUNDATION Copyright 2013 by Immune Deficiency Foundation, USA. REPRINT 2015 Readers may redistribute this article to other individuals for non-commercial use, provided that the text, html codes, and this notice remain intact and unaltered in any way. The Immune Deficiency Foundation Patient & Family Handbook may not be resold, reprinted or redistributed for compensation of any kind without prior written permission from the Immune Deficiency Foundation. If you have any questions about permission, please contact: Immune Deficiency Foundation, 110 West Road, Suite 300, Towson, MD 21204, USA; or by telephone at 800-296-4433. Immune Deficiency Foundation Patient & Family Handbook for Primary Immunodeficency Diseases 5th Edition This publication has been made possible through a generous grant from Baxalta Incorporated Immune Deficiency Foundation 110 West Road, Suite 300 Towson, MD 21204 800-296-4433 www.primaryimmune.org [email protected] EDITORS R. Michael Blaese, MD, Executive Editor Francisco A. Bonilla, MD, PhD Immune Deficiency Foundation Boston Children’s Hospital Towson, MD Boston, MA E. Richard Stiehm, MD M. Elizabeth Younger, CPNP, PhD University of California Los Angeles Johns Hopkins Los Angeles, CA Baltimore, MD CONTRIBUTORS Mark Ballow, MD Joseph Bellanti, MD R. Michael Blaese, MD William Blouin, MSN, ARNP, CPNP State University of New York Georgetown University Hospital Immune Deficiency Foundation Miami Children’s Hospital Buffalo, NY Washington, DC Towson, MD Miami, FL Francisco A. -

Two Locus Inheritance of Non-Syndromic Midline Craniosynostosis Via Rare SMAD6 and 4 Common BMP2 Alleles 5 6 Andrew T
1 2 3 Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and 4 common BMP2 alleles 5 6 Andrew T. Timberlake1-3, Jungmin Choi1,2, Samir Zaidi1,2, Qiongshi Lu4, Carol Nelson- 7 Williams1,2, Eric D. Brooks3, Kaya Bilguvar1,5, Irina Tikhonova5, Shrikant Mane1,5, Jenny F. 8 Yang3, Rajendra Sawh-Martinez3, Sarah Persing3, Elizabeth G. Zellner3, Erin Loring1,2,5, Carolyn 9 Chuang3, Amy Galm6, Peter W. Hashim3, Derek M. Steinbacher3, Michael L. DiLuna7, Charles 10 C. Duncan7, Kevin A. Pelphrey8, Hongyu Zhao4, John A. Persing3, Richard P. Lifton1,2,5,9 11 12 1Department of Genetics, Yale University School of Medicine, New Haven, CT, USA 13 2Howard Hughes Medical Institute, Yale University School of Medicine, New Haven, CT, USA 14 3Section of Plastic and Reconstructive Surgery, Department of Surgery, Yale University School of Medicine, New Haven, CT, USA 15 4Department of Biostatistics, Yale University School of Medicine, New Haven, CT, USA 16 5Yale Center for Genome Analysis, New Haven, CT, USA 17 6Craniosynostosis and Positional Plagiocephaly Support, New York, NY, USA 18 7Department of Neurosurgery, Yale University School of Medicine, New Haven, CT, USA 19 8Child Study Center, Yale University School of Medicine, New Haven, CT, USA 20 9The Rockefeller University, New York, NY, USA 21 22 ABSTRACT 23 Premature fusion of the cranial sutures (craniosynostosis), affecting 1 in 2,000 24 newborns, is treated surgically in infancy to prevent adverse neurologic outcomes. To 25 identify mutations contributing to common non-syndromic midline (sagittal and metopic) 26 craniosynostosis, we performed exome sequencing of 132 parent-offspring trios and 59 27 additional probands. -

Type of the Paper (Article
Supplementary Material A Proteomics Study on the Mechanism of Nutmeg-induced Hepatotoxicity Wei Xia 1, †, Zhipeng Cao 1, †, Xiaoyu Zhang 1 and Lina Gao 1,* 1 School of Forensic Medicine, China Medical University, Shenyang 110122, P. R. China; lessen- [email protected] (W.X.); [email protected] (Z.C.); [email protected] (X.Z.) † The authors contributed equally to this work. * Correspondence: [email protected] Figure S1. Table S1. Peptide fraction separation liquid chromatography elution gradient table. Time (min) Flow rate (mL/min) Mobile phase A (%) Mobile phase B (%) 0 1 97 3 10 1 95 5 30 1 80 20 48 1 60 40 50 1 50 50 53 1 30 70 54 1 0 100 1 Table 2. Liquid chromatography elution gradient table. Time (min) Flow rate (nL/min) Mobile phase A (%) Mobile phase B (%) 0 600 94 6 2 600 83 17 82 600 60 40 84 600 50 50 85 600 45 55 90 600 0 100 Table S3. The analysis parameter of Proteome Discoverer 2.2. Item Value Type of Quantification Reporter Quantification (TMT) Enzyme Trypsin Max.Missed Cleavage Sites 2 Precursor Mass Tolerance 10 ppm Fragment Mass Tolerance 0.02 Da Dynamic Modification Oxidation/+15.995 Da (M) and TMT /+229.163 Da (K,Y) N-Terminal Modification Acetyl/+42.011 Da (N-Terminal) and TMT /+229.163 Da (N-Terminal) Static Modification Carbamidomethyl/+57.021 Da (C) 2 Table S4. The DEPs between the low-dose group and the control group. Protein Gene Fold Change P value Trend mRNA H2-K1 0.380 0.010 down Glutamine synthetase 0.426 0.022 down Annexin Anxa6 0.447 0.032 down mRNA H2-D1 0.467 0.002 down Ribokinase Rbks 0.487 0.000 -

DOCK8 Drives Src-Dependent NK Cell Effector Function Conor J
DOCK8 Drives Src-Dependent NK Cell Effector Function Conor J. Kearney, Stephin J. Vervoort, Kelly M. Ramsbottom, Andrew J. Freeman, Jessica Michie, Jane This information is current as Peake, Jean-Laurent Casanova, Capucine Picard, Stuart G. of October 1, 2021. Tangye, Cindy S. Ma, Ricky W. Johnstone, Katrina L. Randall and Jane Oliaro J Immunol 2017; 199:2118-2127; Prepublished online 9 August 2017; doi: 10.4049/jimmunol.1700751 Downloaded from http://www.jimmunol.org/content/199/6/2118 Supplementary http://www.jimmunol.org/content/suppl/2017/08/09/jimmunol.170075 http://www.jimmunol.org/ Material 1.DCSupplemental References This article cites 38 articles, 12 of which you can access for free at: http://www.jimmunol.org/content/199/6/2118.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on October 1, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2017 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology DOCK8 Drives Src-Dependent NK Cell Effector Function Conor J. -

DOCK8 Regulates Fitness and Function of Regulatory T Cells Through Modulation of IL-2 Signaling
DOCK8 regulates fitness and function of regulatory T cells through modulation of IL-2 signaling Akhilesh K. Singh, … , Troy R. Torgerson, Mohamed Oukka JCI Insight. 2017;2(19):e94275. https://doi.org/10.1172/jci.insight.94275. Research Article Immunology Foxp3+ Tregs possess potent immunosuppressive activity, which is critical for maintaining immune homeostasis and self- tolerance. Defects in Treg development or function result in inadvertent immune activation and autoimmunity. Despite recent advances in Treg biology, we still do not completely understand the molecular and cellular mechanisms governing the development and suppressive function of these cells. Here, we have demonstrated an essential role of the dedicator of cytokinesis 8 (DOCK8), guanine nucleotide exchange factors required for cytoskeleton rearrangement, cell migration, and immune cell survival in controlling Treg fitness and their function. Treg-specific DOCK8 deletion led to spontaneous multiorgan inflammation in mice due to uncontrolled T cell activation and production of proinflammatory cytokines. In addition, we show that DOCK8-deficient Tregs are defective in competitive fitness and in vivo suppressive function. Furthermore, DOCK8 controls IL-2 signaling, crucial for maintenance and competitive fitness of Tregs, via a STAT5- dependent manner. Our study provides potentially novel insights into the essential function of DOCK8 in Tregs and immune regulation, and it explains the autoimmune manifestations associated with DOCK8 deficiency. Find the latest version: https://jci.me/94275/pdf RESEARCH ARTICLE DOCK8 regulates fitness and function of regulatory T cells through modulation of IL-2 signaling Akhilesh K. Singh,1 Ahmet Eken,1 David Hagin,1 Khushbu Komal,1 Gauri Bhise,1 Azima Shaji,1 Tanvi Arkatkar,1 Shaun W. -

Bioinformatics Analysis of Potential Key Genes and Mechanisms in Type 2 Diabetes Mellitus Basavaraj Vastrad1, Chanabasayya Vastrad*2
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.28.437386; this version posted March 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Bioinformatics analysis of potential key genes and mechanisms in type 2 diabetes mellitus Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.03.28.437386; this version posted March 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Type 2 diabetes mellitus (T2DM) is etiologically related to metabolic disorder. The aim of our study was to screen out candidate genes of T2DM and to elucidate the underlying molecular mechanisms by bioinformatics methods. Expression profiling by high throughput sequencing data of GSE154126 was downloaded from Gene Expression Omnibus (GEO) database. The differentially expressed genes (DEGs) between T2DM and normal control were identified. And then, functional enrichment analyses of gene ontology (GO) and REACTOME pathway analysis was performed. Protein–protein interaction (PPI) network and module analyses were performed based on the DEGs. Additionally, potential miRNAs of hub genes were predicted by miRNet database . Transcription factors (TFs) of hub genes were detected by NetworkAnalyst database. Further, validations were performed by receiver operating characteristic curve (ROC) analysis and real-time polymerase chain reaction (RT-PCR). -

Identification of Transcriptional Mechanisms Downstream of Nf1 Gene Defeciency in Malignant Peripheral Nerve Sheath Tumors Daochun Sun Wayne State University
Wayne State University DigitalCommons@WayneState Wayne State University Dissertations 1-1-2012 Identification of transcriptional mechanisms downstream of nf1 gene defeciency in malignant peripheral nerve sheath tumors Daochun Sun Wayne State University, Follow this and additional works at: http://digitalcommons.wayne.edu/oa_dissertations Recommended Citation Sun, Daochun, "Identification of transcriptional mechanisms downstream of nf1 gene defeciency in malignant peripheral nerve sheath tumors" (2012). Wayne State University Dissertations. Paper 558. This Open Access Dissertation is brought to you for free and open access by DigitalCommons@WayneState. It has been accepted for inclusion in Wayne State University Dissertations by an authorized administrator of DigitalCommons@WayneState. IDENTIFICATION OF TRANSCRIPTIONAL MECHANISMS DOWNSTREAM OF NF1 GENE DEFECIENCY IN MALIGNANT PERIPHERAL NERVE SHEATH TUMORS by DAOCHUN SUN DISSERTATION Submitted to the Graduate School of Wayne State University, Detroit, Michigan in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY 2012 MAJOR: MOLECULAR BIOLOGY AND GENETICS Approved by: _______________________________________ Advisor Date _______________________________________ _______________________________________ _______________________________________ © COPYRIGHT BY DAOCHUN SUN 2012 All Rights Reserved DEDICATION This work is dedicated to my parents and my wife Ze Zheng for their continuous support and understanding during the years of my education. I could not achieve my goal without them. ii ACKNOWLEDGMENTS I would like to express tremendous appreciation to my mentor, Dr. Michael Tainsky. His guidance and encouragement throughout this project made this dissertation come true. I would also like to thank my committee members, Dr. Raymond Mattingly and Dr. John Reiners Jr. for their sustained attention to this project during the monthly NF1 group meetings and committee meetings, Dr. -

Patient & Family Handbook
Immune Deficiency Foundation Patient & Family Handbook For Primary Immunodeficiency Diseases This book contains general medical information which cannot be applied safely to any individual case. Medical knowledge and practice can change rapidly. Therefore, this book should not be used as a substitute for professional medical advice. SIXTH EDITION COPYRIGHT 1987, 1993, 2001, 2007, 2013, 2019 IMMUNE DEFICIENCY FOUNDATION Copyright 2019 by Immune Deficiency Foundation, USA. Readers may redistribute this article to other individuals for non-commercial use, provided that the text, html codes, and this notice remain intact and unaltered in any way. The Immune Deficiency Foundation Patient & Family Handbook may not be resold, reprinted or redistributed for compensation of any kind without prior written permission from the Immune Deficiency Foundation. If you have any questions about permission, please contact: Immune Deficiency Foundation, 110 West Road, Suite 300, Towson, MD 21204, USA; or by telephone at 800-296-4433. Immune Deficiency Foundation Patient & Family Handbook For Primary Immunodeficiency Diseases 6th Edition The development of this publication was supported by Shire, now Takeda. 110 West Road, Suite 300 Towson, MD 21204 800.296.4433 www.primaryimmune.org [email protected] Editors Mark Ballow, MD Jennifer Heimall, MD Elena Perez, MD, PhD M. Elizabeth Younger, Executive Editor Children’s Hospital of Philadelphia Allergy Associates of the CRNP, PhD University of South Florida Palm Beaches Johns Hopkins University Jennifer Leiding, -

Hyper Ige Syndromes and DOCK8 Forms of HIES Table 20:1
Immune Deficiency Foundation Patient & Family Handbook For Primary Immunodeficiency Diseases Chapter 20 Immune Deficiency Foundation Patient & Family Handbook For Primary Immunodeficiency Diseases 6th Edition The development of this publication was supported by Shire, now Takeda. 110 West Road, Suite 300 Towson, MD 21204 800.296.4433 www.primaryimmune.org [email protected] Chapter 20 Hyper IgE Syndromes (HIES): STAT3 Loss of Function, DOCK8 Deficiency and Others Alexandra Freeman, MD, National Institutes of Health, Bethesda, Maryland, USA Jennifer Heimall, MD, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA Hyper IgE Syndromes (HIES) are rare forms of primary immunodeficiency diseases (PI) characterized by recurrent eczema, skin abscesses, lung infections, eosinophilia (high numbers of eosinophils in the blood), and high serum levels of immunoglobulin E (IgE). Although initially described as two forms, with autosomal dominant (AD) and autosomal recessive (AR) inheritance, we now recognize that these are two distinct diseases caused by different genetic causes, with the two most common being from harmful mutations in STAT3 causing loss of function (STAT3-LOF) and DOCK8. These diseases share overlapping clinical and laboratory features; however, they also exhibit distinct clinical symptoms, disease courses, and outcomes. In addition, several other genetic variants have since been described to present with similar symptoms. History Clinical Presentation STAT3-LOF was described first as Job Syndrome STAT3 Deficiency in 1966 in two girls with many episodes of STAT3 Deficiency, is associated with heterozygous pneumonia, eczema-like rashes, and recurrent skin loss of function mutations in the transcription factor boils. These boils were remarkable for their lack STAT3. This is the more common form of HIES in of surrounding warmth, redness or tenderness, the U.S. -
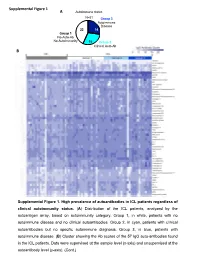
Supplemental Figures (1-6) Unhighlighted
Supplemental Figure 1 A Autoimmune status N=51 Group 3 Autoimmune Disease 22 14 Group 1 No Auto-Ab No Autoimmunity 15 Group 2 Clinical Auto-Ab B Supplemental Figure 1. High prevalence of autoantibodies in ICL patients regardless of clinical autoimmunity status. (A) Distribution of the ICL patients, analyzed by the autoantigen array, based on autoimmunity category. Group 1, in white, patients with no autoimmune disease and no clinical autoantibodies. Group 2, in cyan, patients with clinical autoantibodies but no specific autoimmune diagnosis. Group 3, in blue, patients with autoimmune disease. (B) Cluster showing the Ab scores of the 57 IgG auto-antibodies found in the ICL patients. Data were supervised at the sample level (x-axis) and unsupervised at the autoantibody level (y-axis). (Cont.) Supplemental Figure 1C (Cont.) Supplemental Figure 1. High prevalence of Supplemental Figure 1D autoantibodies in ICL patients regardless of clinical autoimmunity status. (C) Cluster showing the Ab scores of the 39 IgM auto-antibodies found in the ICL patients. Data were supervised at the sample level (x-axis) and unsupervised at the autoantibody level (y-axis). (D) Principal Component Analysis using the combined IgG and IgM Ab scores for each patient. Each symbol represents one patient. ICL patients cluster (blue circle) separately from HC cluster (black discontinuous circle), and the three ICL subgroups cluster together. Pairwise Adonis test with adjusted p values for FDR: p<0.001 for all three comparisons, HC vs group 1, HC vs group 2 and HC vs group 3. Differences within the three ICL subgroups by autoimmunity status were not statistically significant. -

DOCK8 Regulates Signal Transduction Events to Control Immunity
Cellular & Molecular Immunology (2017) 14, 406–411 & 2017 CSI and USTC All rights reserved 2042-0226/17 $32.00 www.nature.com/cmi MINI REVIEW DOCK8 regulates signal transduction events to control immunity Conor J Kearney1,2, Katrina L Randall3,4 and Jane Oliaro1,2 Genetic mutations in the gene encoding DOCK8 cause an autosomal recessive form of hyper immunoglobulin E syndrome (AR-HIES), referred to as DOCK8 deficiency. DOCK8 deficiency in humans results in the onset of combined immunodeficiency disease (CID), clinically associated with chronic infections with diverse microbial pathogens, and a predisposition to malignancy. It is now becoming clear that DOCK8 regulates the function of diverse immune cell sub-types, particularly lymphocytes, to drive both innate and adaptive immune responses. Early studies demonstrated that DOCK8 is required for lymphocyte survival, migration and immune synapse formation, which translates to poor pathogen control in the absence of DOCK8. However, more recent advances have pointed to a crucial role for DOCK8 in regulating the signal transduction events that control transcriptional activity, cytokine production and functional polarization of immune cells. Here, we summarize recent advances in our understanding of DOCK8 function, paying particular attention to an emerging role as a signaling intermediate to promote immune responses to diverse external stimuli. Cellular & Molecular Immunology (2017) 14, 406–411; doi:10.1038/cmi.2017.9; published online 3 April 2017 Keywords: DOCK8; immunodeficiency; lymphocytes; -

270 Genes Genetic Insights Panel
OVER 270 GENES GENETIC INSIGHTS PANEL Dravet Syndrome SCN1A EPILEPSY, Early Infantile epileptic encephalopathy 7 KCNQ2 Early infantile epileptic encephalopathy 11 / Benign familial infantile seizures 3 SCN2A SEIZURES, & OTHER Early infantile epileptic encephalopathy 13 / Benign familial infantile seizures 5 SCN8A NEUROMUSCULAR Ethylmalonic Encephalopathy ETHE1 Familial Infantile Convulsions with Paroxysmal Choreoathetosis PRRT2 CONDITIONS Pyridoxine-Dependent Epilepsy ALDH7A1 Pyridoxal 5'-Phosphate-Dependent Epilepsy PNPO Tuberous Sclerosis Complex (TSC) TSC1, TSC2 Clouston syndrome GJB6 Deafness, Autosomal Dominant, 12 TECTA Deafness, Autosomal Recessive, 6 (DFNB6) TMIE Deafness, Autosomal Recessive, 8 (DFNB8) TMPRSS3 Deafness, Autosomal Recessive, 28 TRIOBP Deafness, Autosomal Recessive, 31 (DFNB31) WHRN Deafness, Autosomal Recessive, 79 TPRN GJB2-Related Hearing Loss GJB2 Hermansky-Pudlak syndrome HPS4 Hermansky-Pudlak Syndrome, Type 1 HPS1 Jervell and Lange-Nielson Syndrome KCNE1 Nonsyndromic Hearing Loss CDH23 HEARING & Ocular Albinism Type I GPR143 Oculocutaneous Albinism Type IV SLC45A2 VISION LOSS Optic Atrophy Type 1 OPA1 Ornithine Aminotransferase Deficiency OAT Pendred Syndrome SLC26A4 Sensorineural Hearing Loss MYO15A Shah-Waardenburg Syndrome SOX10 Usher syndrome type 1C USH1C Usher Syndrome, Type ID CDH23 Usher syndrome type 1G USH1G Usher Syndrome, Type IIA USH2A Usher syndrome type IID WHRN Waardenburg Syndrome PAX3 Alagille Syndrome 1 / Tetralogy of Fallot JAG1 Arrhythmogenic Right Ventricular Cardiomyopathy DSC2 Arrhythmogenic