(Genus: Bombus): from the Organism to the Environment AD
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Conceptual Design Documentation
Appendix A: Conceptual Design Documentation APPENDIX A Conceptual Design Documentation June 2019 A-1 APPENDIX A: CONCEPTUAL DESIGN DOCUMENTATION The environmental analyses in the NEPA and CEQA documents for the proposed improvements at Oceano County Airport (the Airport) are based on conceptual designs prepared to provide a realistic basis for assessing their environmental consequences. 1. Widen runway from 50 to 60 feet 2. Widen Taxiways A, A-1, A-2, A-3, and A-4 from 20 to 25 feet 3. Relocate segmented circle and wind cone 4. Installation of taxiway edge lighting 5. Installation of hold position signage 6. Installation of a new electrical vault and connections 7. Installation of a pollution control facility (wash rack) CIVIL ENGINEERING CALCULATIONS The purpose of this conceptual design effort is to identify the amount of impervious surface, grading (cut and fill) and drainage implications of the projects identified above. The conceptual design calculations detailed in the following figures indicate that Projects 1 and 2, widening the runways and taxiways would increase the total amount of impervious surface on the Airport by 32,016 square feet, or 0.73 acres; a 6.6 percent increase in the Airport’s impervious surface area. Drainage patterns would remain the same as both the runway and taxiways would continue to sheet flow from their centerlines to the edge of pavement and then into open, grassed areas. The existing drainage system is able to accommodate the modest increase in stormwater runoff that would occur, particularly as soil conditions on the Airport are conducive to infiltration. Figure A-1 shows the locations of the seven projects incorporated in the Proposed Action. -
Floral Resource Competition Between Honey Bees and Wild Bees: Is There Clear Evidence and Can We Guide Management and Conservation?
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/325627761 Floral Resource Competition Between Honey Bees and Wild Bees: Is There Clear Evidence and Can We Guide Management and Conservation? Article in Environmental Entomology · June 2018 DOI: 10.1093/ee/nvy077 CITATIONS READS 12 603 4 authors, including: Victoria Wojcik Pollinator Partnership 11 PUBLICATIONS 72 CITATIONS SEE PROFILE All content following this page was uploaded by Victoria Wojcik on 19 June 2018. The user has requested enhancement of the downloaded file. Environmental Entomology, XX(X), 2018, 1–12 doi: 10.1093/ee/nvy077 Pollinator Ecology and Management Review Floral Resource Competition Between Honey Bees and Wild Bees: Is There Clear Evidence and Can We Guide Management and Conservation? Victoria A. Wojcik,1 Lora A. Morandin, Laurie Davies Adams, and Kelly E. Rourke Pollinator Partnership, 423 Washington Street, 5th floor, San Francisco, CA 94111, and 1Corresponding author, e-mail: [email protected] Subject Editor: Gloria DeGrandi-Hoffman Received 31 October 2017; Editorial decision 2 May 2018 Abstract Supporting managed honey bees by pasturing in natural landscapes has come under review due to concerns that honey bees could negatively impact the survival of wild bees through competition for floral resources. Critique and assessment of the existing body of published literature against our criteria focussing on studies that can support best management resulted in 19 experimental papers. Indirect measures of competition examining foraging patterns and behavior yielded equivocal results. Direct measures of reproduction and growth were investigated in only seven studies, with six indicating negative impacts to wild bees from the presence of managed honey bees. -

Bumble Bee Surveys in the Columbia River Gorge National Scenic Area of Oregon and Washington
Bumble Bee Surveys in the Columbia River Gorge National Scenic Area of Oregon and Washington Final report from the Xerces Society to the U.S. Forest Service and Interagency Special Status/Sensitive Species Program (ISSSSP) Agreement L13AC00102, Modification 5 Bombus vosnesenskii on Balsamorhiza sagittata. Photo by Rich Hatfield, the Xerces Society. By Rich Hatfield, Sarina Jepsen, and Scott Black, the Xerces Society for Invertebrate Conservation September 2017 1 Table of Contents Abstract ......................................................................................................................................................... 3 Introduction .................................................................................................................................................. 3 Methods ........................................................................................................................................................ 6 Site Selection ............................................................................................................................................. 6 Site Descriptions (west to east) ................................................................................................................ 7 T14ES27 (USFS) ..................................................................................................................................... 7 Cape Horn (USFS) ................................................................................................................................. -
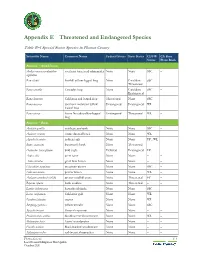
Appendix E Threatened and Endangered Species
Appendix E Threatened and Endangered Species Table E–1 Special Status Species in Plumas County Scientific Name Common Name Federal Status State Status CDFW CA Rare Status Plant Rank Animals – Amphibians Ambystoma macrodactylum southern long-toed salamander None None SSC – sigillatum Rana boylii foothill yellow-legged frog None Candidate SSC – Threatened Rana cascadae Cascades frog None Candidate SSC – Endangered Rana draytonii California red-legged frog Threatened None SSC – Rana muscosa southern mountain yellow- Endangered Endangered WL – legged frog Rana sierrae Sierra Nevada yellow-legged Endangered Threatened WL – frog Animals – Birds Accipiter gentilis northern goshawk None None SSC – Accipiter striatus sharp-shinned hawk None None WL – Aquila chrysaetos golden eagle None None FP ; WL – Buteo swainsoni Swainson's hawk None Threatened – – Haliaeetus leucocephalus bald eagle Delisted Endangered FP – Ardea alba great egret None None – – Ardea herodias great blue heron None None – – Charadrius montanus mountain plover None None SSC – Falco mexicanus prairie falcon None None WL – Antigone canadensis tabida greater sandhill crane None Threatened FP – Riparia riparia bank swallow None Threatened – – Lanius ludovicianus loggerhead shrike None None SSC – Larus californicus California gull None None WL – Pandion haliaetus osprey None None WL – Setophaga petechia yellow warbler None None SSC – Spizella breweri Brewer's sparrow None None – – Phalacrocorax auritus double-crested cormorant None None WL – Melanerpes lewis Lewis' woodpecker None -

Ukiah Western Hills Open Land Acquisition & Limited Development
Ukiah Western Hills Open Land Acquisition & Limited Development Agreement Draft Initial Study & Mitigated Negative Declaration Attachments April 16, 2021 ATTACHMENT A Existing Site Photographs Existing access road Existing water tank site Existing "house site" on one of the proposed Development Parcels ATTACHMENT B Prepared For: Michelle Irace, Planning Manager Department of Community Development 300 Seminary Avenue, Ukiah, CA 95482 APNs: 001-040-83, 157- 070-01, 157-070-02, and 003-190-01 Prepared by Jacobszoon & Associates, Inc. Alicia Ives Ringstad Senior Wildlife Biologist [email protected] Date: March 11, 2021 Updated: April 8, 2021 Biological Assessment Report Table of Contents Section 1.0: Introduction .................................................................................................................................................................. 2 Section 2.0: Regulations and Descriptions ....................................................................................................................................... 2 2.1 Regulatory Setting ................................................................................................................................................................. 2 2.2 Natural Communities and Sensitive Natural Communities .................................................................................................... 3 2.3 Special-Status Species........................................................................................................................................................... -

Assessing the Potential for Deep Learning and Computer Vision to Identify Bumble Bee Species from Images Brian J
www.nature.com/scientificreports OPEN Assessing the potential for deep learning and computer vision to identify bumble bee species from images Brian J. Spiesman1*, Claudio Gratton2, Richard G. Hatfeld3, William H. Hsu4, Sarina Jepsen3, Brian McCornack1, Krushi Patel5 & Guanghui Wang5,6 Pollinators are undergoing a global decline. Although vital to pollinator conservation and ecological research, species-level identifcation is expensive, time consuming, and requires specialized taxonomic training. However, deep learning and computer vision are providing ways to open this methodological bottleneck through automated identifcation from images. Focusing on bumble bees, we compare four convolutional neural network classifcation models to evaluate prediction speed, accuracy, and the potential of this technology for automated bee identifcation. We gathered over 89,000 images of bumble bees, representing 36 species in North America, to train the ResNet, Wide ResNet, InceptionV3, and MnasNet models. Among these models, InceptionV3 presented a good balance of accuracy (91.6%) and average speed (3.34 ms). Species-level error rates were generally smaller for species represented by more training images. However, error rates also depended on the level of morphological variability among individuals within a species and similarity to other species. Continued development of this technology for automatic species identifcation and monitoring has the potential to be transformative for the felds of ecology and conservation. To this end, we present BeeMachine, a web application that allows anyone to use our classifcation model to identify bumble bees in their own images. Bees (Hymenoptera: Anthophila) serve a critical role in most terrestrial ecosystems as pollinators of crops and natural plant communities e.g.,1–3. -

Guide to Bumble Bees of the Western United States
Bumble Bees of the Western United States By Jonathan Koch James Strange A product of the U.S. Forest Service and the Pollinator Partnership Paul Williams with funding from the National Fish and Wildlife Foundation Executive Editor Larry Stritch, Ph.D., USDA Forest Service Cover: Bombus huntii foraging. Photo Leah Lewis Executive and Managing Editor Laurie Davies Adams, The Pollinator Partnership Graphic Design and Art Direction Marguerite Meyer Administration Jennifer Tsang, The Pollinator Partnership IT Production Support Elizabeth Sellers, USGS Alphabetical Quick Reference to Species B. appositus .............110 B. frigidus ..................46 B. rufocinctus ............86 B. balteatus ................22 B. griseocollis ............90 B. sitkensis ................38 B. bifarius ..................78 B. huntii ....................66 B. suckleyi ...............134 B. californicus ..........114 B. insularis ...............126 B. sylvicola .................70 B. caliginosus .............26 B. melanopygus .........62 B. ternarius ................54 B. centralis ................34 B. mixtus ...................58 B. terricola ...............106 B. crotchii ..................82 B. morrisoni ...............94 B. vagans ...................50 B. fernaldae .............130 B. nevadensis .............18 B. vandykei ................30 B. fervidus................118 B. occidentalis .........102 B. vosnesenskii ..........74 B. flavifrons ...............42 B. pensylvanicus subsp. sonorus ....122 B. franklini .................98 2 Bumble Bees of the -

IUCN Assessments for North American Bombus Spp
IUCN Assessments for North American Bombus spp. Prepared by: Rich Hatfield*±, Sheila Colla†, Sarina Jepsen*, Leif Richardson‡, Robbin Thorp∆, and Sarah Foltz Jordan* Assessments completed December 2014 Document updated March 2, 2015 Bombus occidentalis on Solidago canadensis. Photo by R. Hatfield ± Corresponding author: [email protected] * The Xerces Society for Invertebrate Conservation, 628 NE Broadway, Suite 200, Portland, OR 97232, xerces.org † Wildlife Preservation Canada 5420 Side Road 6, Guelph, ON N1H 6J2 CANADA wildlifepreservation.ca ‡ Gund Institute for Ecological Economics, University of Vermont, 617 Main Street Burlington, VT 05405 ∆ University of California at Davis, Department of Entomology and Nematology Main Office, UC Davis Briggs Hall, Room 367, Davis, CA 95616-5270 Table of Contents Introducon ............................................................................................................................................................. 3 Methods ................................................................................................................................................................... 4 Bombus affinis .......................................................................................................................................................... 8 Bombus appositus .................................................................................................................................................... 9 Bombus auricomus .................................................................................................................................................. -

Measuring and Monitoring Pollinator Diversity Along Elevation Gradients in Olympic and North Cascades National Parks
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science Measuring and Monitoring Pollinator Diversity along Elevation Gradients in Olympic and North Cascades National Parks Natural Resource Report NPS/NOCA/NRR—2018/1634 ON THE COVER Row one (left to right): Bombus on Arnica (NOCA), subalpine meadow on McAlester Trail (NOCA); Row two: bee bowl on Obstruction Point Rd. (OLYM), B. occidentalis on thistle (Cirsium; OLYM); Row three: Fireweed (Chamerion) above Ross Lake (NOCA), J. Freilich and J.D. Herndon collecting bees in OLYM All photos by Jessica Rykken Measuring and Monitoring Pollinator Diversity along Elevation Gradients in Olympic and North Cascades National Parks Natural Resource Report NPS/NOCA/NRR—2018/1634 Jessica J. Rykken Museum of Comparative Zoology Harvard University 26 Oxford Street, Cambridge, MA 02138 May 2018 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. The series supports the advancement of science, informed decision-making, and the achievement of the National Park Service mission. The series also provides a forum for presenting more lengthy results that may not be accepted by publications with page limitations. -

General Biological Resources Assessment and Jurisdictional Delineation for the Big Bear Solar Observatory Solis Telescope Project
General Biological Resources Assessment And Jurisdictional Delineation For the Big Bear Solar Observatory Solis Telescope Project City of Big Bear San Bernardino County, California Prepared for: Transtech Engineers, Inc. Attn: Bob Carlisle 577 Bonanza Trail, Suite B Big Bear Lake, CA 92315 Prepared January 2017 Prepared by: Jericho Systems, Inc. Shay Lawrey, President 47 1st Street Redlands, CA 92373 Certification Jericho Systems, Inc. 47 1st Street Redlands, CA 92373 (909) 915-5900 Contact: Shay Lawrey, President and Ecologist/Regulatory Specialist Certification: I hereby certify that the statements furnished herein, and in the attached exhibits present data and information required for this Biological Resources Repot to the best of my ability, and the facts, statements, and information presented are true and correct to the best of my knowledge and belief. This report was prepared in accordance with professional requirements and standards. Fieldwork conducted for this assessment was performed by me. I certify that I have not signed a non-disclosure or consultant confidentiality agreement with the project proponent and that I have no financial interest in the project. ______________________________________ Shay Lawrey, Ecologist/Regulatory Specialist Big Bear Solar Observatory JERICHO SYSTEMS, INC. Solis Telescope Project BRA & JD ii Table of Contents CERTIFICATION .......................................................................................................................................... II 1 INTRODUCTION .................................................................................................................................. -

Appendix N Biological Database Queries and List of Species Observed
APPENDIX N BIOLOGICAL DATABASE QUERIES AND LIST OF SPECIES OBSERVED NATURAL RESOURCES CONSERVATION SERVICE REPORT United States A product of the National Custom Soil Resource Department of Cooperative Soil Survey, Agriculture a joint effort of the United Report for States Department of Agriculture and other Shasta-Trinity National Federal agencies, State Natural agencies including the Forest Area, Parts of Resources Agricultural Experiment Conservation Stations, and local Humboldt, Siskiyou, Shasta, Service participants Tehama, and Trinity Counties, California; and Siskiyou County, California, Central Part September 6, 2016 Preface Soil surveys contain information that affects land use planning in survey areas. They highlight soil limitations that affect various land uses and provide information about the properties of the soils in the survey areas. Soil surveys are designed for many different users, including farmers, ranchers, foresters, agronomists, urban planners, community officials, engineers, developers, builders, and home buyers. Also, conservationists, teachers, students, and specialists in recreation, waste disposal, and pollution control can use the surveys to help them understand, protect, or enhance the environment. Various land use regulations of Federal, State, and local governments may impose special restrictions on land use or land treatment. Soil surveys identify soil properties that are used in making various land use or land treatment decisions. The information is intended to help the land users identify and reduce the effects of soil limitations on various land uses. The landowner or user is responsible for identifying and complying with existing laws and regulations. Although soil survey information can be used for general farm, local, and wider area planning, onsite investigation is needed to supplement this information in some cases. -
Appendix 3 Biological Resources Assessment
Initial Study Maple Hill Community Fields Project – Conditional Use Permit APN: 0312-311-13 and 0312-311-25 March 2021 APPENDIX 3 BIOLOGICAL RESOURCES ASSESSMENT BIOLOGICAL RESOURCES ASSESSMENT FOR THE BEAR VALLEY UNIFIED SCHOOL DISTRICT EDUCATION FOUNDATION MAPLE HILL FIELDS COMPLEX PROJECT Unincorporated Community of Sugarloaf, San Bernardino County, California USGS – Big Bear City Quadrangle Section 13 of Township 2 North, Range 1 East Date Prepared: February 2021 Prepared for: Bear Valley Unified School District Education Foundation PO Box 1529 Big Bear Lake, CA 92315 On Behalf of: Tom Dodson and Associates 2150 N Arrowhead Avenue San Bernardino, CA 92405 Prepared by: Jacobs Engineering Group, Inc. 55616 Pipes Canyon Road Yucca Valley, California 92284 CERTIFICATION I certify under penalty of law that I have personally examined and am familiar with the information submitted in this document and all attachments and that, based on my inquiry of those individuals immediately responsible for obtaining the information, I believe that the information is true, accurate, and complete. I am aware that there are significant penalties for submitting false information, including the possibility of fine and imprisonment. _________________________________________________________________________ Lisa Patterson, National Senior Environmental Project Manager 2021 Tom Dodson & Associates BVUSD Education Foundation Maple Hill Fields Complex Project Biological Resources Assessment ii Table of Contents 1 INTRODUCTION ..................................................................................................................................