Supplemental Materials 1 SUPPLEMENTAL METHODS CSC
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transcriptional Control of Tissue-Resident Memory T Cell Generation
Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2019 © 2019 Filip Cvetkovski All rights reserved ABSTRACT Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Tissue-resident memory T cells (TRM) are a non-circulating subset of memory that are maintained at sites of pathogen entry and mediate optimal protection against reinfection. Lung TRM can be generated in response to respiratory infection or vaccination, however, the molecular pathways involved in CD4+TRM establishment have not been defined. Here, we performed transcriptional profiling of influenza-specific lung CD4+TRM following influenza infection to identify pathways implicated in CD4+TRM generation and homeostasis. Lung CD4+TRM displayed a unique transcriptional profile distinct from spleen memory, including up-regulation of a gene network induced by the transcription factor IRF4, a known regulator of effector T cell differentiation. In addition, the gene expression profile of lung CD4+TRM was enriched in gene sets previously described in tissue-resident regulatory T cells. Up-regulation of immunomodulatory molecules such as CTLA-4, PD-1, and ICOS, suggested a potential regulatory role for CD4+TRM in tissues. Using loss-of-function genetic experiments in mice, we demonstrate that IRF4 is required for the generation of lung-localized pathogen-specific effector CD4+T cells during acute influenza infection. Influenza-specific IRF4−/− T cells failed to fully express CD44, and maintained high levels of CD62L compared to wild type, suggesting a defect in complete differentiation into lung-tropic effector T cells. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -

SUPPLEMENTARY TABLES and FIGURE LEGENDS Supplementary
SUPPLEMENTARY TABLES AND FIGURE LEGENDS Supplementary Figure 1. Quantitation of MYC levels in vivo and in vitro. a) MYC levels in cell lines 6814, 6816, 5720, 966, and 6780 (corresponding to first half of Figure 1a in main text). MYC is normalized to tubulin. b) MYC quantitations (normalized to tubulin) for cell lines Daudi, Raji, Jujoye, KRA, KRB, GM, and 6780 corresponding to second half of Figure 1a. c) In vivo MYC quantitations, for mice treated with 0-0.5 ug/ml doxycycline in their drinking water. MYC is normalized to tubulin. d) Quantitation of changing MYC levels during in vitro titration, normalized to tubulin. e) Levels of Odc (normalized to tubulin) follow MYC levels in titration series. Supplementary Figure 2. Evaluation of doxycycline concentration in the plasma of mice treated with doxycycline in their drinking water. Luciferase expressing CHO cells (Tet- off) (Clonethech Inc) that is responsive to doxycycline by turning off luciferase expression was treated with different concentrations of doxycycline in culture. A standard curve (blue line) correlating luciferase activity (y-axis) with treatment of doxycycline (x- axis) was generated for the CHO cell in culture. Plasma from mice treated with different concentrations of doxycycline in their drinking water was separated and added to the media of the CHO cells. Luciferase activity was measured and plotted on the standard curve (see legend box). The actual concentration of doxycycline in the plasma was extrapolated for the luciferase activity measured. The doxycycline concentration 0.2 ng/ml measured in the plasma of mice correlates with 0.05 μg/ml doxycycline treatment in the drinking water of mice, the in vivo threshold for tumor regression. -

Profilin-1 Is Required for Survival of Adult Hematopoietic Stem Cells
Extended methods Immunohistochemistry HepG-2, SMMC-7721, and 293T cells were obtained from Cell Resource Center of Shanghai Institute for Biological Science, Chinese Academy Science, Shanghai, China. HUVEC cells were kindly provided by Prof. Ping-Jin Gao at Institute of Health Sciences (Shanghai, China). All these cell lines were cultured in DMEM with 10% FBS. MDA- MB-231 cell line was kindly provided by Prof. Ming-Yao Liu (East China Normal University, Shanghai, China) and was cultured in Leibovitz L-15 medium with 10% FBS. All these cell lines were originally purchased from ATCC. MDA-MB-231, SMMC-7721 or HepG2 cells were grown on coverslips in 24-well plates and fixed in either 4% paraformaldehyde or pre-chilled methanol (-20°C) for 10 min. In some cases, WT or VPS33B-null Lin-Sca-1+c-Kit+Flk2-CD34- LT-HSCs were collected by flow cytometry and fixed for immunofluorescence staining. Cells were then blocked with 3% BSA in PBS for 60 min followed by incubation with primary antibodies overnight. The antibodies used were anti-HA (Sigma), anti-Flag (Sigma), anti-VPS33B (Sigma), anti- VPS16B (Abcam), anti-GDI2 (Proteintech), anti-LAMP1 (Proteintech), anti-FLOT1 (Abways), anti-CD63 (Proteintech), anti-ANGPTL2 (R&D system), anti-ANGPTL3 (R&D system), anti-TPO (Abways), anti-GLUT1 (Proteintech), anti-LDHA (Proteintech), anti-PKM2 (CST), anti-RAB11A (Abways), anti-RAB27A (Abways) and anti-V5 (Biodragon). Fluorescent-conjugated secondary antibodies (Alexa Fluor® 488 or Alexa Fluor® 555) against mouse, rabbit, or goat were obtained from the Thermo Scientific Inc. The details for all the antibodies are listed in Table S3. -

Cell Communication and Signaling Biomed Central
Cell Communication and Signaling BioMed Central Review Open Access Extravasation of leukocytes in comparison to tumor cells Carina Strell and Frank Entschladen* Address: Institute of Immunology, Witten/Herdecke University, Stockumer Str. 10, 58448 Witten, Germany Email: Carina Strell - [email protected]; Frank Entschladen* - [email protected] * Corresponding author Published: 4 December 2008 Received: 18 November 2008 Accepted: 4 December 2008 Cell Communication and Signaling 2008, 6:10 doi:10.1186/1478-811X-6-10 This article is available from: http://www.biosignaling.com/content/6/1/10 © 2008 Strell and Entschladen; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract The multi-step process of the emigration of cells from the blood stream through the vascular endothelium into the tissue has been termed extravasation. The extravasation of leukocytes is fairly well characterized down to the molecular level, and has been reviewed in several aspects. Comparatively little is known about the extravasation of tumor cells, which is part of the hematogenic metastasis formation. Although the steps of the process are basically the same in leukocytes and tumor cells, i.e. rolling, adhesion, transmigration (diapedesis), the molecules that are involved are different. A further important difference is that leukocyte interaction with the endothelium changes the endothelial integrity only temporarily, whereas tumor cell interaction leads to an irreversible damage of the endothelial architecture. Moreover, tumor cells utilize leukocytes for their extravasation as linkers to the endothelium. -
Download Validation Data
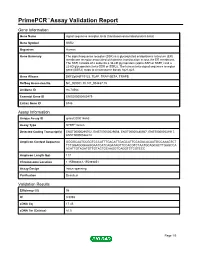
PrimePCR™Assay Validation Report Gene Information Gene Name signal sequence receptor, beta (translocon-associated protein beta) Gene Symbol SSR2 Organism Human Gene Summary The signal sequence receptor (SSR) is a glycosylated endoplasmic reticulum (ER) membrane receptor associated with protein translocation across the ER membrane. The SSR consists of 2 subunits a 34-kD glycoprotein (alpha-SSR or SSR1) and a 22-kD glycoprotein (beta-SSR or SSR2). The human beta-signal sequence receptor gene (SSR2) maps to chromosome bands 1q21-q23. Gene Aliases DKFZp686F19123, TLAP, TRAP-BETA, TRAPB RefSeq Accession No. NC_000001.10, NT_004487.19 UniGene ID Hs.74564 Ensembl Gene ID ENSG00000163479 Entrez Gene ID 6746 Assay Information Unique Assay ID qHsaCID0014663 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000295702, ENST00000529008, ENST00000480567, ENST00000531917, ENST00000526212 Amplicon Context Sequence GGGGCAATCCGGTCCCATTTGACATTGAGCATTCCAGACACAATGCCAAAGTCT TCTGGAGGGAAGGAATCATCAGATAGTTCCACGTCTAATGCAGCACTTGAGCCA ACATTGTAGATGTTGTACTGCAAGGTCAGGTCTCGTCCC Amplicon Length (bp) 117 Chromosome Location 1:155988061-155989851 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9998 cDNA Cq 17.45 cDNA Tm (Celsius) 81.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report SSR2, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

Lysosomal Membrane Glycoproteins Bind Cholesterol and Contribute to Lysosomal Cholesterol Export
1 Lysosomal membrane glycoproteins bind cholesterol and 2 contribute to lysosomal cholesterol export 3 4 Jian Li and Suzanne R. Pfeffer* 5 Department of Biochemistry, Stanford University School of Medicine 6 Stanford, CA USA 94305-5307 7 8 *Correspondence to: [email protected]. 9 10 Abstract: LAMP1 and LAMP2 proteins are highly abundant, ubiquitous, mammalian proteins 11 that line the lysosome limiting membrane, and protect it from lysosomal hydrolase action. 12 LAMP2 deficiency causes Danon’s disease, an X-linked hypertrophic cardiomyopathy. LAMP2 13 is needed for chaperone-mediated autophagy, and its expression improves tissue function in 14 models of aging. We show here that LAMP1 and LAMP2 bind cholesterol in a manner that 15 buries the cholesterol 3β-hydroxyl group; they also bind tightly to NPC1 and NPC2 proteins that 16 export cholesterol from lysosomes. Quantitation of cellular LAMP2 and NPC1 protein levels 17 suggest that LAMP proteins represent a significant cholesterol binding site at the lysosome 18 limiting membrane, and may signal cholesterol availability. Functional rescue experiments show 19 that the ability of LAMP2 to facilitate cholesterol export from lysosomes relies on its ability to 20 bind cholesterol directly. 21 22 23 Introduction 24 Eukaryotic lysosomes are acidic, membrane-bound organelles that contain proteases, lipases and 25 nucleases and degrade cellular components to regenerate catabolic precursors for cellular use (1- 26 3). Lysosomes are crucial for the degradation of substrates from the cytoplasm, as well as 27 membrane bound compartments derived from the secretory, endocytic, autophagic and 28 phagocytic pathways. The limiting membrane of lysosomes is lined with so-called lysosomal 29 membrane glycoproteins (LAMPs) that are comprised of a short cytoplasmic domain, a single 30 transmembrane span, and a highly, N- and O-glycosylated lumenal domain (4-6). -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

A Crosstalk Between the RNA Binding Protein Smaug and the Hedgehog Pathway Links Cell Signaling to Mrna Regulation in Drosophila Lucía Bruzzone
A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila Lucía Bruzzone To cite this version: Lucía Bruzzone. A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila. Cellular Biology. Université Sorbonne Paris Cité, 2018. English. NNT : 2018USPCC234. tel-02899776 HAL Id: tel-02899776 https://tel.archives-ouvertes.fr/tel-02899776 Submitted on 15 Jul 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Thèse de doctorat de l’Université Sorbonne Paris Cité Préparée à l’Université Paris Diderot Ecole doctorale HOB n° 561 Institut Jacques Monod / Equipe Développement, Signalisation et Trafic A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in Drosophila Lucía Bruzzone Thèse de doctorat de Biologie Dirigée par Anne Plessis Présentée et soutenue publiquement à Paris le 19 mars 2018 Président du jury: Alain Zider / Professeur Université Paris Diderot -

Disease-Related Cellular Protein Networks Differentially Affected
www.nature.com/scientificreports OPEN Disease‑related cellular protein networks diferentially afected under diferent EGFR mutations in lung adenocarcinoma Toshihide Nishimura1,8*, Haruhiko Nakamura1,2,8, Ayako Yachie3,8, Takeshi Hase3,8, Kiyonaga Fujii1,8, Hirotaka Koizumi4, Saeko Naruki4, Masayuki Takagi4, Yukiko Matsuoka3, Naoki Furuya5, Harubumi Kato6,7 & Hisashi Saji2 It is unclear how epidermal growth factor receptor EGFR major driver mutations (L858R or Ex19del) afect downstream molecular networks and pathways. This study aimed to provide information on the infuences of these mutations. The study assessed 36 protein expression profles of lung adenocarcinoma (Ex19del, nine; L858R, nine; no Ex19del/L858R, 18). Weighted gene co-expression network analysis together with analysis of variance-based screening identifed 13 co-expressed modules and their eigen proteins. Pathway enrichment analysis for the Ex19del mutation demonstrated involvement of SUMOylation, epithelial and mesenchymal transition, ERK/mitogen- activated protein kinase signalling via phosphorylation and Hippo signalling. Additionally, analysis for the L858R mutation identifed various pathways related to cancer cell survival and death. With regard to the Ex19del mutation, ROCK, RPS6KA1, ARF1, IL2RA and several ErbB pathways were upregulated, whereas AURK and GSKIP were downregulated. With regard to the L858R mutation, RB1, TSC22D3 and DOCK1 were downregulated, whereas various networks, including VEGFA, were moderately upregulated. In all mutation types, CD80/CD86 (B7), MHC, CIITA and IFGN were activated, whereas CD37 and SAFB were inhibited. Costimulatory immune-checkpoint pathways by B7/CD28 were mainly activated, whereas those by PD-1/PD-L1 were inhibited. Our fndings may help identify potential therapeutic targets and develop therapeutic strategies to improve patient outcomes. -

Sensitization to the Lysosomal Cell Death Pathway by Oncogene- Induced Down-Regulation of Lysosome-Associated Membrane Proteins 1 and 2
Research Article Sensitization to the Lysosomal Cell Death Pathway by Oncogene- Induced Down-regulation of Lysosome-Associated Membrane Proteins 1 and 2 Nicole Fehrenbacher,1 Lone Bastholm,2 Thomas Kirkegaard-Sørensen,1 Bo Rafn,1 Trine Bøttzauw,1 Christina Nielsen,1 Ekkehard Weber,3 Senji Shirasawa,4 Tuula Kallunki,1 and Marja Ja¨a¨ttela¨1 1Apoptosis Department and Centre for Genotoxic Stress Response, Institute for Cancer Biology, Danish Cancer Society; 2Institute of Molecular Pathology, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark; 3Institute of Physiological Chemistry, Medical Faculty, Martin-Luther-University Halle-Wittenberg, Halle, Germany; and 4Department of Cell Biology, School of Medicine, Fukuoka University, Fukuoka, Japan Abstract molecules of the cell to breakdown products available for Expression and activity of lysosomal cysteine cathepsins metabolic reuse (3, 4). Cathepsin proteases are among the best- correlate with the metastatic capacity and aggressiveness of studied lysosomal hydrolases. They are maximally active at the tumors. Here, we show that transformation of murine acidic pH of lysosomes (pH 4–5). However, many of them can be Y527F embryonic fibroblasts with v-H-ras or c-src changes the active at the neutral pH outside lysosomes, albeit with a decreased distribution, density, and ultrastructure of the lysosomes, efficacy and/or altered specificity (5). For example, transformation and tumor environment enhance the expression of lysosomal decreases the levels of lysosome-associated membrane pro- teins (LAMP-1 and LAMP-2) in an extracellular signal- cysteine cathepsins and increase their secretion into the extracel- regulated kinase (ERK)- and cathepsin-dependent manner, lular space (6). Once outside the tumor cells, cathepsins stimulate and sensitizes the cells to lysosomal cell death pathways angiogenesis, tumor growth, and invasion in murine cancer induced by various anticancer drugs (i.e., cisplatin, etoposide, models, thereby enhancing cancer progression (7, 8).