Analyzing Huge Pathology Images with Open Source Software
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Panstamps Documentation Release V0.5.3
panstamps Documentation Release v0.5.3 Dave Young 2020 Getting Started 1 Installation 3 1.1 Troubleshooting on Mac OSX......................................3 1.2 Development...............................................3 1.2.1 Sublime Snippets........................................4 1.3 Issues...................................................4 2 Command-Line Usage 5 3 Documentation 7 4 Command-Line Tutorial 9 4.1 Command-Line..............................................9 4.1.1 JPEGS.............................................. 12 4.1.2 Temporal Constraints (Useful for Moving Objects)...................... 17 4.2 Importing to Your Own Python Script.................................. 18 5 Installation 19 5.1 Troubleshooting on Mac OSX...................................... 19 5.2 Development............................................... 19 5.2.1 Sublime Snippets........................................ 20 5.3 Issues................................................... 20 6 Command-Line Usage 21 7 Documentation 23 8 Command-Line Tutorial 25 8.1 Command-Line.............................................. 25 8.1.1 JPEGS.............................................. 28 8.1.2 Temporal Constraints (Useful for Moving Objects)...................... 33 8.2 Importing to Your Own Python Script.................................. 34 8.2.1 Subpackages.......................................... 35 8.2.1.1 panstamps.commonutils (subpackage)........................ 35 8.2.1.2 panstamps.image (subpackage)............................ 35 8.2.2 Classes............................................ -

Discrete Cosine Transform for 8X8 Blocks with CUDA
Discrete Cosine Transform for 8x8 Blocks with CUDA Anton Obukhov [email protected] Alexander Kharlamov [email protected] October 2008 Document Change History Version Date Responsible Reason for Change 0.8 24.03.2008 Alexander Kharlamov Initial release 0.9 25.03.2008 Anton Obukhov Added algorithm-specific parts, fixed some issues 1.0 17.10.2008 Anton Obukhov Revised document structure October 2008 2 Abstract In this whitepaper the Discrete Cosine Transform (DCT) is discussed. The two-dimensional variation of the transform that operates on 8x8 blocks (DCT8x8) is widely used in image and video coding because it exhibits high signal decorrelation rates and can be easily implemented on the majority of contemporary computing architectures. The key feature of the DCT8x8 is that any pair of 8x8 blocks can be processed independently. This makes possible fully parallel implementation of DCT8x8 by definition. Most of CPU-based implementations of DCT8x8 are firmly adjusted for operating using fixed point arithmetic but still appear to be rather costly as soon as blocks are processed in the sequential order by the single ALU. Performing DCT8x8 computation on GPU using NVIDIA CUDA technology gives significant performance boost even compared to a modern CPU. The proposed approach is accompanied with the sample code “DCT8x8” in the NVIDIA CUDA SDK. October 2008 3 1. Introduction The Discrete Cosine Transform (DCT) is a Fourier-like transform, which was first proposed by Ahmed et al . (1974). While the Fourier Transform represents a signal as the mixture of sines and cosines, the Cosine Transform performs only the cosine-series expansion. -

Oracle FLEXCUBE Core Banking Licensing Guide Release 11.5.0.0.0
Oracle FLEXCUBE Core Banking Licensing Guide Release 11.5.0.0.0 Part No. E52876-01 July 2014 Licensing Guide July 2014 Oracle Financial Services Software Limited Oracle Park Off Western Express Highway Goregaon (East) Mumbai, Maharashtra 400 063 India Worldwide Inquiries: Phone: +91 22 6718 3000 Fax:+91 22 6718 3001 www.oracle.com/financialservices/ Copyright © 2008, 2014, Oracle and/or its affiliates. All rights reserved. Oracle and Java are registered trademarks of Oracle and/or its affiliates. Other names may be trademarks of their respective owners. U.S. GOVERNMENT END USERS: Oracle programs, including any operating system, integrated software, any programs installed on the hardware, and/or documentation, delivered to U.S. Government end users are “commercial computer software” pursuant to the applicable Federal Acquisition Regulation and agency-specific supplemental regulations. As such, use, duplication, disclosure, modification, and adaptation of the programs, including any operating system, integrated software, any programs installed on the hardware, and/or documentation, shall be subject to license terms and license restrictions applicable to the programs. No other rights are granted to the U.S. Government. This software or hardware is developed for general use in a variety of information management applications. It is not developed or intended for use in any inherently dangerous applications, including applications that may create a risk of personal injury. If you use this software or hardware in dangerous applications, then you shall be responsible to take all appropriate failsafe, backup, redundancy, and other measures to ensure its safe use. Oracle Corporation and its affiliates disclaim any liability for any damages caused by use of this software or hardware in dangerous applications. -

Java Advanced Imaging
Java Advanced Imaging MIMUC Medientechnik SS 2003 Übersicht Wozu Bildbearbeitung mit Frameworks? Warum JAI? Verzahnung mit Java2D (AWT) JAI Packages und Typen Unterstützte Codecs und Bildformate BufferedImage, Raster, ColorModel (AWT) Rendergraphen Aufbau der Bildklassen Anzeigen eines gespeicherten Bildes Parameterblock Operationen Look-Up-Table Speichern eines Bildes Quellangabe MIMUC Medientechnik SS 2003 Helge Groß 2 - 23 Wozu Bildbearbeitung mit Frameworks? Rechner/Monitore können Bilder heute schnell verarbeiten/anzeigen Dadurch ist Analyse- und Bearbeitungsmöglichkeit am PC geschaffen Frameworks stellen bereit Bildmanipulationsmöglichkeiten Mathematische Operationen Bildanzeigemöglichkeit Bild-Codecs Einsatzgebiete Bildanalyse- und Bildanzeigesysteme Objektpositionierung, Objekttracking, Qualitätsprüfung Bildbearbeitungssysteme Medien, Werbeindustrie, Grafik-Design, Zeichenprogramme Applikationen Programme, Computerspiele World Wide Web Webseiten MIMUC Medientechnik SS 2003 Helge Groß 3 - 23 Warum JAI? Pro Plattformunabhängig Geringer Speicherplatz Schnelle Internetübertragung Applets im WWW Bildverarbeitende Web-Applikationen Als Frontend einer anderen Programmiersprache Diese übernimmt komplexere Operationen Client-Server-Architektur Komplexe Bildmanipulationen durchaus praktikabel Großer Leistungsumfang zur Bildverarbeitung Versucht alle Bedürfnisse zu erfüllen Extrem Erweiterbar Contra Langsam nicht zuletzt wegen Swing MIMUC Medientechnik SS 2003 Helge Groß 4 - 23 Verzahnung mit Java2D (AWT) JAI -

RISC-V Software Ecosystem
RISC-V Software Ecosystem Palmer Dabbelt [email protected] UC Berkeley February 8, 2015 Software on RISC-V So it turns out there is a lot of software... 2 Software on RISC-V sys-libs/zlib-1.2.8-r1 media-libs/libjpeg-turbo-1.3.1-r1 virtual/shadow-0 virtual/libintl-0-r1 sys-apps/coreutils-8.23 sys-apps/less-471 sys-libs/ncurses-5.9-r3 sys-libs/readline-6.3 p8-r2 app-admin/eselect-python-20140125 sys-apps/gentoo-functions-0.8 sys-libs/glibc-2.20-r1 sys-apps/grep-2.21-r1 dev-libs/gmp-6.0.0a sys-apps/util-linux-2.25.2-r2 virtual/service-manager-0 sys-libs/db-6.0.30-r1 sys-apps/sed-4.2.2 virtual/editor-0 virtual/libiconv-0-r1 sys-apps/file-5.22 sys-devel/gcc-4.9.2-r1 app-arch/bzip2-1.0.6-r7 dev-libs/mpfr-3.1.2 p10 x11-libs/libX11-1.6.2 sys-apps/busybox-1.23.0-r1 sys-process/psmisc-22.21-r2 virtual/pager-0 sys-devel/gcc-config-1.8 net-misc/netifrc-0.3.1 x11-libs/libXext-1.3.3 sys-libs/timezone-data-2014j dev-libs/popt-1.16-r2 x11-libs/libXfixes-5.0.1 app-misc/editor-wrapper-4 sys-devel/binutils-config-4-r1 x11-libs/libXt-1.1.4 net-firewall/iptables-1.4.21-r1 virtual/libffi-3.0.13-r1 x11-libs/fltk-1.3.3-r2 sys-libs/e2fsprogs-libs-1.42.12 sys-libs/cracklib-2.9.2 x11-libs/libXi-1.7.4 dev-libs/libpipeline-1.4.0 sys-apps/kmod-19 x11-libs/libXtst-1.2.2 sys-libs/gdbm-1.11 sys-devel/make-4.1-r1 net-misc/tigervnc-1.3.1-r2 app-portage/portage-utils-0.53 sys-process/procps-3.3.10-r1 dev-lang/perl-5.20.1-r4 sys-apps/sandbox-2.6-r1 sys-apps/iproute2-3.18.0 app-admin/perl-cleaner-2.19 app-misc/pax-utils-0.9.2 virtual/dev-manager-0 perl-core/Data-Dumper-2.154.0 -

I Deb, You Deb, Everybody Debs: Debian Packaging For
I deb, you deb, everybody debs Debian packaging for beginners and experts alike Ondřej Surý • [email protected] • [email protected] • 25. 10. 2017 Contents ● .deb binary package structure ● Source package structure ● Basic toolchain ● Recommended toolchain ● Keeping the sources in git ● Clean build environment ● Misc... ● So how do I become Debian Developer? My Debian portfolio (since 2000) ● Mostly team maintained ● BIRD ● PHP + PECL (pkg-php) ● Cyrus SASL ○ Co-installable packages since 7.x ● Cyrus IMAPD ○ Longest serving PHP maintainer in Debian ● Apache2 + mod_md (fresh) (and still not crazy) ● ...other little stuff ● libjpeg-turbo ○ Transitioned Debian from IIJ JPEG (that Older work crazy guy) to libjpeg-turbo ● DNS Packaging Group ● GTK/GNOME/Freedesktop ○ CZ.NIC’s Knot DNS and Knot Resolver ● Redmine/Ruby ○ NLnet Lab’s NSD, Unbound, getdns, ldns Never again, it’s a straight road to madness ○ PowerDNS ○ ○ BIND 9 ● Berkeley DB and LMDB (pkg-db) ○ One Berkeley DB per release (yay!) Binary package structure ● ar archive consisting of: $ ar xv knot_2.0.1-4_amd64.deb x – debian-binary ○ debian-binary x – control.tar.gz x – data.tar.xz ■ .deb format version (2.0) ○ control.tar.gz $ dpkg-deb -X knot_2.0.1-4_amd64.deb output/ ./ ■ Package informatio (control) ./etc/ ■ Maintainer scripts […] ./usr/sbin/knotd ● {pre,post}{inst,rm} […] Misc (md5sum, conffiles) ■ $ dpkg-deb -e knot_2.0.1-4_amd64.deb DEBIAN/ ○ data.tar.xz $ ls DEBIAN/ conffiles control md5sums postinst postrm preinst ■ Actual content of the package prerm ■ This is what gets installed $ dpkg -I knot_2.0.1-4_amd64.deb ● Nástroje pro práci s .deb soubory new debian package, version 2.0. -
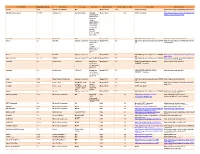
Third Party Version
Third Party Name Third Party Version Manufacturer License Type Comments Merge Product Merge Product Versions License details Software source autofac 3.5.2 Autofac Contributors MIT Merge Cardio 10.2 SOUP repository https://www.nuget.org/packages/Autofac/3.5 .2 Gibraltar Loupe Agent 2.5.2.815 eSymmetrix Gibraltor EULA Gibraltar Merge Cardio 10.2 SOUP repository https://my.gibraltarsoftware.com/Support/Gi Loupe Agent braltar_2_5_2_815_Download will be used within the Cardio Application to view events and metrics so you can resolve support issues quickly and easily. Modernizr 2.8.3 Modernizr MIT Merge Cadio 6.0 http://modernizr.com/license/ http://modernizr.com/download/ drools 2.1 Red Hat Apache License 2.0 it is a very old Merge PACS 7.0 http://www.apache.org/licenses/LICENSE- http://mvnrepository.com/artifact/drools/dro version of 2.0 ols-spring/2.1 drools. Current version is 6.2 and license type is changed too drools 6.3 Red Hat Apache License 2.0 Merge PACS 7.1 http://www.apache.org/licenses/LICENSE- https://github.com/droolsjbpm/drools/releases/ta 2.0 g/6.3.0.Final HornetQ 2.2.13 v2.2..13 JBOSS Apache License 2.0 part of JBOSS Merge PACS 7.0 http://www.apache.org/licenses/LICENSE- http://mvnrepository.com/artifact/org.hornet 2.0 q/hornetq-core/2.2.13.Final jcalendar 1.0 toedter.com LGPL v2.1 MergePacs Merge PACS 7.0 GNU LESSER GENERAL PUBLIC http://toedter.com/jcalendar/ server uses LICENSE Version 2. v1, and viewer uses v1.3. -

An Optimization of JPEG-LS Using an Efficient and Low-Complexity
Received April 26th, 2021. Revised June 27th, 2021. Accepted July 23th, 2021. Digital Object Identifier 10.1109/ACCESS.2021.3100747 LOCO-ANS: An Optimization of JPEG-LS Using an Efficient and Low-Complexity Coder Based on ANS TOBÍAS ALONSO , GUSTAVO SUTTER , AND JORGE E. LÓPEZ DE VERGARA High Performance Computing and Networking Research Group, Escuela Politécnica Superior, Universidad Autónoma de Madrid, Spain. {tobias.alonso, gustavo.sutter, jorge.lopez_vergara}@uam.es This work was supported in part by the Spanish Research Agency under the project AgileMon (AEI PID2019-104451RB-C21). ABSTRACT Near-lossless compression is a generalization of lossless compression, where the codec user is able to set the maximum absolute difference (the error tolerance) between the values of an original pixel and the decoded one. This enables higher compression ratios, while still allowing the control of the bounds of the quantization errors in the space domain. This feature makes them attractive for applications where a high degree of certainty is required. The JPEG-LS lossless and near-lossless image compression standard combines a good compression ratio with a low computational complexity, which makes it very suitable for scenarios with strong restrictions, common in embedded systems. However, our analysis shows great coding efficiency improvement potential, especially for lower entropy distributions, more common in near-lossless. In this work, we propose enhancements to the JPEG-LS standard, aimed at improving its coding efficiency at a low computational overhead, particularly for hardware implementations. The main contribution is a low complexity and efficient coder, based on Tabled Asymmetric Numeral Systems (tANS), well suited for a wide range of entropy sources and with simple hardware implementation. -

Programming in Java Advanced Imaging
Programming in Java™ Advanced Imaging Release 1.0.1 November 1999 JavaSoft A Sun Microsystems, Inc. Business 901 San Antonio Road Palo Alto, CA 94303 USA 415 960-1300 fax 415 969-9131 1999 Sun Microsystems, Inc. 901 San Antonio Road, Palo Alto, California 94303 U.S.A. All rights reserved. RESTRICTED RIGHTS LEGEND: Use, duplication, or disclosure by the United States Government is subject to the restrictions set forth in DFARS 252.227-7013 (c)(1)(ii) and FAR 52.227-19. The release described in this document may be protected by one or more U.S. patents, for- eign patents, or pending applications. Sun Microsystems, Inc. (SUN) hereby grants to you a fully paid, nonexclusive, nontrans- ferable, perpetual, worldwide limited license (without the right to sublicense) under SUN’s intellectual property rights that are essential to practice this specification. This license allows and is limited to the creation and distribution of clean-room implementa- tions of this specification that (i) are complete implementations of this specification, (ii) pass all test suites relating to this specification that are available from SUN, (iii) do not derive from SUN source code or binary materials, and (iv) do not include any SUN binary materials without an appropriate and separate license from SUN. Java, JavaScript, Java 3D, and Java Advanced Imaging are trademarks of Sun Microsys- tems, Inc. Sun, Sun Microsystems, the Sun logo, Java and HotJava are trademarks or reg- istered trademarks of Sun Microsystems, Inc. UNIX® is a registered trademark in the United States and other countries, exclusively licensed through X/Open Company, Ltd. -

CES Free Or Open Source Licenses Licenses Library Version 15.2 2020R1
CES Free or Open Source Licenses Licenses Library Version 15.2 2020R1 Revision 2.00 December 2020 Verint.com Twitter.com/verint Facebook.com/verint Blog.verint.com Table of Contents Free or Open Source Licenses ....................................................................................... 1 7-Zip - GNU LGPL + unRAR restrictions .................................................................... 1 ActivePython ............................................................................................................... 2 ANTLR .......................................................................................................................... 6 Apache License............................................................................................................ 6 ares Library................................................................................................................. 11 Attribution-NonCommercial-ShareAlike 3.0 Unported ............................................. 12 Batik SVG Toolkit ....................................................................................................... 17 Bouncy Castle ............................................................................................................ 19 Boost ........................................................................................................................... 20 BSD (4-Clause) License ............................................................................................ 20 COMMON DEVELOPMENT AND DISTRIBUTION -

LIBJPEG the Independent JPEG Group's JPEG Software README for Release 6B of 27-Mar-1998 This Distribution Contains the Sixth
LIBJPEG The Independent JPEG Group's JPEG software README for release 6b of 27-Mar-1998 This distribution contains the sixth public release of the Independent JPEG Group's free JPEG software. You are welcome to redistribute this software and to use it for any purpose, subject to the conditions under LEGAL ISSUES, below. Serious users of this software (particularly those incorporating it into larger programs) should contact IJG at [email protected] to be added to our electronic mailing list. Mailing list members are notified of updates and have a chance to participate in technical discussions, etc. This software is the work of Tom Lane, Philip Gladstone, Jim Boucher, Lee Crocker, Julian Minguillon, Luis Ortiz, George Phillips, Davide Rossi, Guido Vollbeding, Ge' Weijers, and other members of the Independent JPEG Group. IJG is not affiliated with the official ISO JPEG standards committee. DOCUMENTATION ROADMAP This file contains the following sections: OVERVIEW General description of JPEG and the IJG software. LEGAL ISSUES Copyright, lack of warranty, terms of distribution. REFERENCES Where to learn more about JPEG. ARCHIVE LOCATIONS Where to find newer versions of this software. RELATED SOFTWARE Other stuff you should get. FILE FORMAT WARS Software *not* to get. TO DO Plans for future IJG releases. Other documentation files in the distribution are: User documentation: install.doc How to configure and install the IJG software. usage.doc Usage instructions for cjpeg, djpeg, jpegtran, rdjpgcom, and wrjpgcom. *.1 Unix-style man pages for programs (same info as usage.doc). wizard.doc Advanced usage instructions for JPEG wizards only. -

Compiling Your Own Perl
APPENDIX A Compiling Your Own Perl Compiling Perl on a Unix-like system is simple. First, obtain the source for Perl from CPAN (dppl6++_l]j*lanh*knc+on_+NA=@IA*dpih). Then input the following sequence of commands: p]nvtrblanh)1*4*4*p]n*cv _`lanh)1*4*4 od?kjbecqna)`ao i]ga i]gapaop oq`ki]gaejop]hh On most Unix systems, this code will result in your lanh being installed into the +qon+ hk_]h+ directory tree. If you want it installed elsewhere—for example, in the local directory in your home directory—then replace od?kjbecqna)`a with the following: od?kjbecqna)`ao)@lnabet9z+hk_]h+ which should enable you to install Perl on your computer without root access. Note that the )`ao flag uses all the default options for compiling Perl. If you know that you want nonstandard configuration, just use the flag )`a instead to be prompted for your requirements. Be aware that the source for Perl 5.10.0 requires a patch to work properly with Catalyst. This is fixed in subsequent versions of Perl 5.10. If you need to test code guaranteed to run on a wide range of systems, you should con- sider using Perl version 5.8.7. Perl versions greater than 5.8.7 contain features that were not available in earlier versions of Perl, so Perl 5.8.7 is feature complete for all versions of Perl that Catalyst will run on (version 5.8.1 and later). Put another way, versions 5.8.8 and later have new features that you can’t rely on in earlier releases.