Identification of Promising Prognostic Genes for Relapsed Acute
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -

Transcriptomic Profiling of Equine and Viral Genes in Peripheral Blood
pathogens Article Transcriptomic Profiling of Equine and Viral Genes in Peripheral Blood Mononuclear Cells in Horses during Equine Herpesvirus 1 Infection Lila M. Zarski 1, Patty Sue D. Weber 2, Yao Lee 1 and Gisela Soboll Hussey 1,* 1 Department of Pathobiology and Diagnostic Investigation, Michigan State University, East Lansing, MI 48824, USA; [email protected] (L.M.Z.); [email protected] (Y.L.) 2 Department of Large Animal Clinical Sciences, Michigan State University, East Lansing, MI 48824, USA; [email protected] * Correspondence: [email protected] Abstract: Equine herpesvirus 1 (EHV-1) affects horses worldwide and causes respiratory dis- ease, abortions, and equine herpesvirus myeloencephalopathy (EHM). Following infection, a cell- associated viremia is established in the peripheral blood mononuclear cells (PBMCs). This viremia is essential for transport of EHV-1 to secondary infection sites where subsequent immunopathol- ogy results in diseases such as abortion or EHM. Because of the central role of PBMCs in EHV-1 pathogenesis, our goal was to establish a gene expression analysis of host and equine herpesvirus genes during EHV-1 viremia using RNA sequencing. When comparing transcriptomes of PBMCs during peak viremia to those prior to EHV-1 infection, we found 51 differentially expressed equine genes (48 upregulated and 3 downregulated). After gene ontology analysis, processes such as the interferon defense response, response to chemokines, the complement protein activation cascade, cell adhesion, and coagulation were overrepresented during viremia. Additionally, transcripts for EHV-1, EHV-2, and EHV-5 were identified in pre- and post-EHV-1-infection samples. Looking at Citation: Zarski, L.M.; Weber, P.S.D.; micro RNAs (miRNAs), 278 known equine miRNAs and 855 potentially novel equine miRNAs were Lee, Y.; Soboll Hussey, G. -

Transposable Elements Resistant to Epigenetic Resetting in the Human Germline Are Epigenetic Hotspots for Development and Disease
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.19.998930; this version posted March 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Transposable elements resistant to epigenetic resetting in the human germline are epigenetic hotspots for development and disease Sabine Dietmann1,2,3,11*, Michael J Keogh4,5,11, Walfred Tang2, Erna Magnusdottir6, Toshihiro Kobayashi7,8, Patrick F Chinnery5,9 and M. Azim Surani 1,2,10* ** 1. Wellcome Trust – Medical Research Council Stem Cell Institute, University of Cambridge, UK 2. Wellcome Trust/Cancer Research UK Gurdon Institute, University of Cambridge, Cambridge, UK 3. Department of Developmental Biology and Division of Nephrology, Washington University School of Medicine, St. Louis, MO, USA 4. Institute of Genetic Medicine, Newcastle University, Newcastle Upon Tyne, UK 5. MRC-Mitochondrial Biology Unit, University of Cambridge, Cambridge, UK 6. Department of Biomedical Science and Anatomy, Faculty of Medicine, University of Iceland, Reykjavik, Iceland 7. Section of Mammalian Transgenesis, Center for Genetic Analysis of Behavior, National Institute for Physiological Sciences, Okazaki, Japan 8. Department of Physiological Sciences, The Graduate University of Advanced Studies, Okazaki, Japan 9. Department of Clinical Neurosciences, Cambridge Biomedical Campus, University of Cambridge, Cambridge, UK 10. Department of Physiology and Development of Neuroscience, University of Cambridge, UK 11. These authors contributed equally * Co-corresponding authors; [email protected]; [email protected] ** lead contact:[email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.03.19.998930; this version posted March 20, 2020. -

Revostmm Vol 10-4-2018 Ingles Maquetaciûn 1
108 ORIGINALS / Rev Osteoporos Metab Miner. 2018;10(4):108-18 Roca-Ayats N1, Falcó-Mascaró M1, García-Giralt N2, Cozar M1, Abril JF3, Quesada-Gómez JM4, Prieto-Alhambra D5,6, Nogués X2, Mellibovsky L2, Díez-Pérez A2, Grinberg D1, Balcells S1 1 Departamento de Genética, Microbiología y Estadística - Facultad de Biología - Universidad de Barcelona - Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER) - Instituto de Salud Carlos III (ISCIII) - Instituto de Biomedicina de la Universidad de Barcelona (IBUB) - Instituto de Investigación Sant Joan de Déu (IRSJD) - Barcelona (España) 2 Unidad de Investigación en Fisiopatología Ósea y Articular (URFOA); Instituto Hospital del Mar de Investigaciones Médicas (IMIM) - Parque de Salud Mar - Centro de Investigación Biomédica en Red de Fragilidad y Envejecimiento Saludable (CIBERFES); Instituto de Salud Carlos III (ISCIII) - Barcelona (España) 3 Departamento de Genética, Microbiología y Estadística; Facultad de Biología; Universidad de Barcelona - Instituto de Biomedicina de la Universidad de Barcelona (IBUB) - Barcelona (España) 4 Unidad de Metabolismo Mineral; Instituto Maimónides de Investigación Biomédica de Córdoba (IMIBIC); Hospital Universitario Reina Sofía - Centro de Investigación Biomédica en Red de Fragilidad y Envejecimiento Saludable (CIBERFES); Instituto de Salud Carlos III (ISCIII) - Córdoba (España) 5 Grupo de Investigación en Enfermedades Prevalentes del Aparato Locomotor (GREMPAL) - Instituto de Investigación en Atención Primaria (IDIAP) Jordi Gol - Centro de Investigación -

ALPK1 Hotspot Mutation As a Driver of Human Spiradenoma and Spiradenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-019-09979-0 OPEN ALPK1 hotspot mutation as a driver of human spiradenoma and spiradenocarcinoma Mamunur Rashid1,14, Michiel van der Horst2,14, Thomas Mentzel3, Francesca Butera4, Ingrid Ferreira1, Alena Pance 1, Arno Rütten3, Bostjan Luzar5, Zlatko Marusic6, Nicolas de Saint Aubain7, Jennifer S. Ko8, Steven D. Billings8,Sofia Chen1, Marie Abi Daoud9, James Hewinson1, Sandra Louzada1, Paul W. Harms10, Guia Cerretelli11, Carla Daniela Robles-Espinoza1,12, Rajiv M. Patel10, Louise van der Weyden1, Chris Bakal 4, Jason L. Hornick13, Mark J. Arends 11, Thomas Brenn9,11,15 & David J. Adams1,15 1234567890():,; Spiradenoma and cylindroma are distinctive skin adnexal tumors with sweat gland differ- entiation and potential for malignant transformation and aggressive behaviour. We present the genomic analysis of 75 samples from 57 representative patients including 15 cylindromas, 17 spiradenomas, 2 cylindroma–spiradenoma hybrid tumors, and 24 low- and high-grade spiradenocarcinoma cases, together with morphologically benign precursor regions of these cancers. We reveal somatic or germline alterations of the CYLD gene in 15/15 cylindromas and 5/17 spiradenomas, yet only 2/24 spiradenocarcinomas. Notably, we find a recurrent missense mutation in the kinase domain of the ALPK1 gene in spiradenomas and spir- adenocarcinomas, which is mutually exclusive from mutation of CYLD and can activate the NF-κB pathway in reporter assays. In addition, we show that high-grade spiradenocarcinomas carry loss-of-function TP53 mutations, while cylindromas may have disruptive mutations in DNMT3A. Thus, we reveal the genomic landscape of adnexal tumors and therapeutic targets. 1 Experimental Cancer Genetics, Wellcome Trust Sanger Institute, Cambridge CB10 1SA, UK. -

Alpha Kinase 1 Controls Intestinal Infiammation by Suppressing the IL
ARTICLE DOI: 10.1038/s41467-018-06085-5 OPEN Alpha kinase 1 controls intestinal inflammation by suppressing the IL-12/Th1 axis Grigory Ryzhakov1, Nathaniel R. West1,6, Fanny Franchini1, Simon Clare2, Nicholas E. Ilott1, Stephen N. Sansom1, Samuel J. Bullers1, Claire Pearson1, Alice Costain1, Alun Vaughan-Jackson1, Jeremy A. Goettel3, Joerg Ermann 4, Bruce H. Horwitz4, Ludovico Buti5, Xin Lu 5, Subhankar Mukhopadhyay2, Scott B. Snapper3 & Fiona Powrie1 1234567890():,; Inflammatory bowel disease (IBD) are heterogenous disorders of the gastrointestinal tract caused by a spectrum of genetic and environmental factors. In mice, overlapping regions of chromosome 3 have been associated with susceptibility to IBD-like pathology, including a locus called Hiccs. However, the specific gene that controls disease susceptibility remains unknown. Here we identify a Hiccs locus gene, Alpk1 (encoding alpha kinase 1), as a potent regulator of intestinal inflammation. In response to infection with the commensal pathobiont Helicobacter hepaticus (Hh), Alpk1-deficient mice display exacerbated interleukin (IL)-12/IL-23 dependent colitis characterized by an enhanced Th1/interferon(IFN)-γ response. Alpk1 controls intestinal immunity via the hematopoietic system and is highly expressed by mononuclear phagocytes. In response to Hh, Alpk1−/− macrophages produce abnormally high amounts of IL-12, but not IL-23. This study demonstrates that Alpk1 promotes intestinal homoeostasis by regulating the balance of type 1/type 17 immunity following microbial challenge. 1 Kennedy Institute of Rheumatology, University of Oxford, Oxford OX3 7FY, United Kingdom. 2 Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, United Kingdom. 3 Boston Children’s Hospital and Harvard Medical School, Boston, MA 02115, USA. -

Elabscience.Com ® E-Mail:[email protected] Elabscience Elabscience Biotechnology Inc
Produktinformation Diagnostik & molekulare Diagnostik Laborgeräte & Service Zellkultur & Verbrauchsmaterial Forschungsprodukte & Biochemikalien Weitere Information auf den folgenden Seiten! See the following pages for more information! Lieferung & Zahlungsart Lieferung: frei Haus Bestellung auf Rechnung SZABO-SCANDIC Lieferung: € 10,- HandelsgmbH & Co KG Erstbestellung Vorauskassa Quellenstraße 110, A-1100 Wien T. +43(0)1 489 3961-0 Zuschläge F. +43(0)1 489 3961-7 [email protected] • Mindermengenzuschlag www.szabo-scandic.com • Trockeneiszuschlag • Gefahrgutzuschlag linkedin.com/company/szaboscandic • Expressversand facebook.com/szaboscandic Tel:240-252-7368(USA) Fax:240-252-7376(USA) www.elabscience.com ® E-mail:[email protected] Elabscience Elabscience Biotechnology Inc. ALPK1 Polyclonal Antibody Catalog No. E-AB-52176 Reactivity H Storage Store at -20℃. Avoid freeze / thaw cycles. Host Rabbit Applications IHC,ELISA Isotype IgG Note: Centrifuge before opening to ensure complete recovery of vial contents. Images Immunogen Information Immunogen Fusion protein of human ALPK1 Gene Accession BC060780 Swissprot Q96QP1 Synonyms Alpha protein kinase 1,Alpha-protein kinase 1,ALPK1,ALPK1,Chromosome 4 kinase,LAK,Lymphocyte alpha kinase Immunohistochemistry of paraffin- Product Information embedded Human tonsil tissue using Buffer PBS with 0.05% NaN3 and 40% Glycerol,pH7.4 ALPK1 Polyclonal Antibody at Purify Antigen affinity purification dilution of 1:50(×200) Dilution IHC 1:50-1:300, ELISA 1:5000-1:10000 Background This gene encodes an alpha kinase. Mice which were homozygous for disrupted copies of this gene exhibited coordination defects (PMID: Immunohistochemistry of paraffin- 21208416). Multiple transcript variants encoding different isoforms have embedded Human brain tissue using been found for this gene. ALPK1 (Alpha Kinase 1) is a Protein Coding ALPK1 Polyclonal Antibody at gene. -

Quantitative Lymphatic Vessel Trait Analysis Suggests Vcam1 As Candidate Modifier Gene of Inflammatory Bowel Disease
Genes and Immunity (2010) 11, 219–231 & 2010 Macmillan Publishers Limited All rights reserved 1466-4879/10 $32.00 www.nature.com/gene ORIGINAL ARTICLE Quantitative lymphatic vessel trait analysis suggests Vcam1 as candidate modifier gene of inflammatory bowel disease G Jurisic1, JP Sundberg2, A Bleich3, EH Leiter2, KW Broman4, G Buechler3, L Alley2, D Vestweber5 and M Detmar1 1Institute of Pharmaceutical Sciences, Swiss Federal Institute of Technology, ETH Zurich, Zurich, Switzerland; 2The Jackson Laboratory, Bar Harbor, ME, USA; 3Institute for Laboratory Animal Science and Central Animal Facility, Hannover Medical School, Hannover, Germany; 4Department of Biostatistics and Medical Informatics, University of Wisconsin, Madison, WI, USA and 5Max Planck Institute of Molecular Biomedicine, Mu¨nster, Germany Inflammatory bowel disease (IBD) is a chronic debilitating disease resulting from a complex interaction of multiple genetic factors with the environment. To identify modifier genes of IBD, we used an F2 intercross of IBD-resistant C57BL/6J-Il10À/À mice and IBD-susceptible C3H/HeJBir-Il10À/À (C3Bir-Il10À/À) mice. We found a prominent involvement of lymphatic vessels in IBD and applied a scoring system to quantify lymphatic vascular changes. Quantitative trait locus (QTL) analyses revealed a large-effect QTL on chromosome 3, mapping to an interval of 43.6 Mbp. This candidate interval was narrowed by fine mapping to 22 Mbp, and candidate genes were analyzed by a systems genetics approach that included quantitative gene expression profiling, search for functional polymorphisms, and haplotype block analysis. We identified vascular adhesion molecule 1 (Vcam1) as a candidate modifier gene in the interleukin 10-deficient mouse model of IBD. -

MAFB Determines Human Macrophage Anti-Inflammatory
MAFB Determines Human Macrophage Anti-Inflammatory Polarization: Relevance for the Pathogenic Mechanisms Operating in Multicentric Carpotarsal Osteolysis This information is current as of October 4, 2021. Víctor D. Cuevas, Laura Anta, Rafael Samaniego, Emmanuel Orta-Zavalza, Juan Vladimir de la Rosa, Geneviève Baujat, Ángeles Domínguez-Soto, Paloma Sánchez-Mateos, María M. Escribese, Antonio Castrillo, Valérie Cormier-Daire, Miguel A. Vega and Ángel L. Corbí Downloaded from J Immunol 2017; 198:2070-2081; Prepublished online 16 January 2017; doi: 10.4049/jimmunol.1601667 http://www.jimmunol.org/content/198/5/2070 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2017/01/15/jimmunol.160166 Material 7.DCSupplemental References This article cites 69 articles, 22 of which you can access for free at: http://www.jimmunol.org/content/198/5/2070.full#ref-list-1 by guest on October 4, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2017 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. -

Page 1 Exploring the Understudied Human Kinome For
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.02.022277; this version posted June 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Exploring the understudied human kinome for research and therapeutic opportunities Nienke Moret1,2,*, Changchang Liu1,2,*, Benjamin M. Gyori2, John A. Bachman,2, Albert Steppi2, Rahil Taujale3, Liang-Chin Huang3, Clemens Hug2, Matt Berginski1,4,5, Shawn Gomez1,4,5, Natarajan Kannan,1,3 and Peter K. Sorger1,2,† *These authors contributed equally † Corresponding author 1The NIH Understudied Kinome Consortium 2Laboratory of Systems Pharmacology, Department of Systems Biology, Harvard Program in Therapeutic Science, Harvard Medical School, Boston, Massachusetts 02115, USA 3 Institute of Bioinformatics, University of Georgia, Athens, GA, 30602 USA 4 Department of Pharmacology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA 5 Joint Department of Biomedical Engineering at the University of North Carolina at Chapel Hill and North Carolina State University, Chapel Hill, NC 27599, USA Key Words: kinase, human kinome, kinase inhibitors, drug discovery, cancer, cheminformatics, † Peter Sorger Warren Alpert 432 200 Longwood Avenue Harvard Medical School, Boston MA 02115 [email protected] cc: [email protected] 617-432-6901 ORCID Numbers Peter K. Sorger 0000-0002-3364-1838 Nienke Moret 0000-0001-6038-6863 Changchang Liu 0000-0003-4594-4577 Ben Gyori 0000-0001-9439-5346 John Bachman 0000-0001-6095-2466 Albert Steppi 0000-0001-5871-6245 Page 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.04.02.022277; this version posted June 30, 2020. -
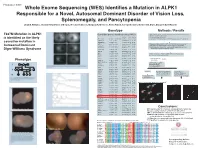
T237M Mutation in ALPK1 Is Identified As the Likely Causative Mutation In
Program #: 3367 Whole Exome Sequencing (WES) Identifies a Mutation in ALPK1 Responsible for a Novel, Autosomal Dominant Disorder of Vision Loss, Splenomegaly, and Pancytopenia Lloyd B. Williams, Chad D. Huff, Denise J. Morgan, Rosann Robinson, Margaux A. Morrison, Krista Kinard, George Rodgers, Kathleen B. Digre, Margaret M. DeAngelis Genotype Methods / Results WES on 4 individuals - 2 affected II-3 III-2, and 2 unaffected II-2 and III-3 T237M Mutation in ALPK1 Candi date genes identified using VAAST Illuimina Truseq Enrichment Kit position Allele Protein Quantitative PCR enrichment is identified as the likely Gene name p-value chromosome (hg19) change change Sequencing with Illumina HiSeq 2000 101 cycle paired end sequencing PRAMEF11 6.10E-06 chr1 12887174 C->T R->H causative mutation in ANKRD20A4 7.32E-06 chr9 69423637 G->A E->K MRPL4 8.55E-06 chr19 10367459 C->T R->W Mapped and aligned with Picard Tools (http://picard.sourceforge.net) FAM90A10 9.77E-06 chr8 7629232 G->T A->S SNPs and indels identified with Genome analysis toolkit (GATK) Autosomal Dominant Manual inspection and curation was done with Intergrative Genomics GOLGA6L10 9.77E-06 chr15 82635194 T->C E->G Viewer (http://www.broadinstitute.org/igv) Digre-Williams Syndrome FAM90A20 1.28E-05 chr8 7155458 C->G A->G EEF1A1 1.65E-05 chr6 74228474 C->T R->H MSI1 1.65E-05 chr12 120800875 C->T V->M VAAST - compares to HGMD, dbSNP, ESP, and1000 Genomes VDAC2 1.71E-05 chr10 76980685 G->T A->S panels to rule out common variants. PIM1 2.08E-05 chr6 37138779 A->T K->M USP11 2.26E-05 chrX -

SUPPLEMENTARY MATERIALS and METHODS PBMC Transcriptomics
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut SUPPLEMENTARY MATERIALS AND METHODS PBMC transcriptomics identifies immune-metabolism disorder during the development of HBV-ACLF Contents l Supplementary methods l Supplementary Figure 1 l Supplementary Figure 2 l Supplementary Figure 3 l Supplementary Figure 4 l Supplementary Figure 5 l Supplementary Table 1 l Supplementary Table 2 l Supplementary Table 3 l Supplementary Table 4 l Supplementary Tables 5-14 l Supplementary Table 15 l Supplementary Table 16 l Supplementary Table 17 Li J, et al. Gut 2021;0:1–13. doi: 10.1136/gutjnl-2020-323395 BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut SUPPLEMENTARY METHODS Test for HBV DNA The levels of HBV DNA were detected using real-time PCR with a COBAS® AmpliPrep/COBAS® TaqMan 48 System (Roche, Basel, Switzerland) and HBV Test v2.0. Criteria for diagnosing cirrhosis Pathology The gold standard for the diagnosis of cirrhosis is a liver biopsy obtained through a percutaneous or transjugular approach.1 Ultrasonography was performed 2-4 hours before biopsy. Liver biopsy specimens were obtained by experienced physicians. Percutaneous transthoracic puncture of the liver was performed according to the standard criteria. After biopsy, patients were monitored in the hospital with periodic analyses of haematocrit and other vital signs for 24 hours. Cirrhosis was diagnosed according to the globally agreed upon criteria.2 Cirrhosis is defined based on its pathological features under a microscope: (a) the presence of parenchymal nodules, (b) differences in liver cell size and appearance, (c) fragmentation of the biopsy specimen, (d) fibrous septa, and (d) an altered architecture and vascular relationships.