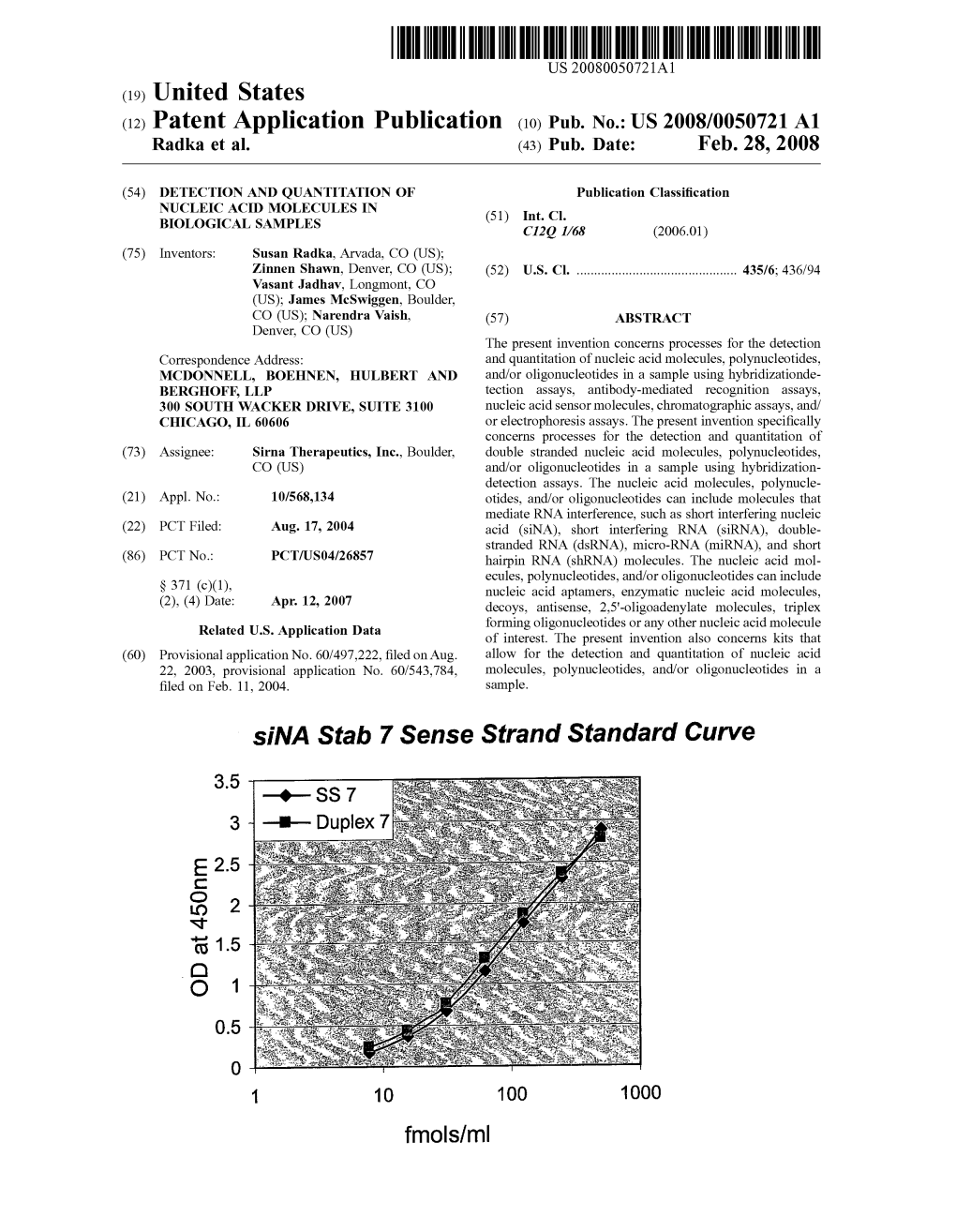
Pub. No.: US 2008/0050721 A1 Sina Stab 7 Sense Strand Standard Curve
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pharmacokinetics, Pharmacodynamics and Metabolism Of
PHARMACOKINETICS, PHARMACODYNAMICS AND METABOLISM OF GTI-2040, A PHOSPHOROTHIOATE OLIGONUCLEOTIDE TARGETING R2 SUBUNIT OF RIBONUCLEOTIDE REDUCTASE DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Xiaohui Wei, M.S. * * * * * * The Ohio State University 2006 Approved by Dissertation Committee: Dr. Kenneth K. Chan, Adviser Adviser Dr. Guido Marcucci, Co-adviser Graduate Program in Pharmacy Dr. Thomas D. Schmittgen Dr. Robert J. Lee Co-Adviser Graduate Program in Pharmacy ABSTRACT Over the last several decades, antisense therapy has been developed into a promising gene-targeted strategy to specifically inhibit the gene expression. Ribonucleotide reductase (RNR), composing of subunits R1 and R2, is an important enzyme involved in the synthesis of all of the precursors used in DNA replication. Over- expression of R2 has been found in almost every type of cancer studied. GTI-2040 is a 20-mer phosphorothioate oligonucleotide targeting the coding region in mRNA of the R2 component of human RNR. In this project, clinical pharamcokinetics (PK), pharmacodynamics (PD) and metabolism of this novel therapeutics were investigated in patients with acute myeloid leukemia (AML). A picomolar specific hybridization-ligation ELISA method has been developed and validated for quantification of GTI-2040. GTI-2040 and neophectin complex was found to enhance drug cellular uptake and exhibited sequence- and dose-dependent down-regulation of R2 mRNA and protein in K562 cells. Robust intracellular concentrations (ICs) of GTI-2040 were achieved in peripheral blood mononuclear cells (PBMC) and bone marrow (BM) cells from treated AML patients. GTI-2040 concentrations in the nucleus of BM cells were found to correlate with the R2 mRNA down-regulation and disease response. -

Whole-Genome Gene Expression Direct Hybridization Assay Guide
Whole-Genome Gene Expression Direct Hybridization Assay Guide For Research Use Only 1,000 0000000 1,00000 100 11000 1,0001,1 ILLUMINA PROPRIETARY Catalog # BD-901-1002 Part # 11322355 Rev. A Notice This document and its contents are proprietary to Illumina, Inc. and its affiliates ("Illumina"), and are intended solely for the contractual use of its customer in connection with the use of the product(s) described herein and for no other purpose. This document and its contents shall not be used or distributed for any other purpose and/or otherwise communicated, disclosed, or reproduced in any way whatsoever without the prior written consent of Illumina. Illumina does not convey any license under its patent, trademark, copyright, or common-law rights nor similar rights of any third parties by this document. The instructions in this document must be strictly and explicitly followed by qualified and properly trained personnel in order to ensure the proper and safe use of the product(s) described herein. All of the contents of this document must be fully read and understood prior to using such product(s). FAILURE TO COMPLETELY READ AND EXPLICITLY FOLLOW ALL OF THE INSTRUCTIONS CONTAINED HEREIN MAY RESULT IN DAMAGE TO THE PRODUCT(S), INJURY TO PERSONS, INCLUDING TO USERS OR OTHERS, AND DAMAGE TO OTHER PROPERTY. ILLUMINA DOES NOT ASSUME ANY LIABILITY ARISING OUT OF THE IMPROPER USE OF THE PRODUCT(S) DESCRIBED HEREIN (INCLUDING PARTS THEREOF OR SOFTWARE) OR ANY USE OF SUCH PRODUCT(S) OUTSIDE THE SCOPE OF THE EXPRESS WRITTEN LICENSES OR PERMISSIONS GRANTED BY ILLUMINA IN CONNECTION WITH CUSTOMER'S ACQUISITION OF SUCH PRODUCT(S). -

Development of Rapid Gene Expression Analysis and Its
ESPOO 2007 VTT PUBLICATIONS 661 VTT PUBLICATIONS 661 Development of rapid gene expression analysis and its application to bioprocess monitoring VTT PUBLICATIONS 661 Development 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 Bioprocesses are used for production of industrial compounds and for 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 studying biological phenomena. In order to be able to better understand 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 and control biological systems, development of methods that generate 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 12345678901234567890123456789012123456789012345678901234567890121234567890123456789012345678901 -
(ERCC) Reference Material Using a Modified Latin Square Design
bioRxiv preprint doi: https://doi.org/10.1101/034868; this version posted January 7, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design Pine PS1, Munro SA1, Parsons JR1, McDaniel J1, Bergstrom Lucas A2, Lozach J3, Myers TG4, Su Q4, Jacobs-Helber SM5,6, and Salit M1 1. Joint Initiative for Metrology in Biology, National Institute of Standards and Technology, Stanford, CA, 94305, USA 2. Genomics Research and Development, Agilent Technologies, Santa Clara, CA, 95051, USA 3. Illumina, Inc., San Diego, CA, 92122, USA 4. National Institute of Allergy and Infectious Diseases, Bethesda, MD, 20892, USA 5. AIBioTech, Inc., Richmond, VA, 23235, USA 6. Current affiliation: GENETWORx, LLC., Glen Allen, VA, USA Corresponding Author: P Scott Pine ([email protected]) Joint Initiative for Metrology in Biology, National Institute of Standards and Technology, 443 Via Ortega, Stanford, CA, 94305, USA E-mail: Sarah A Munro ([email protected]) Jerod R Parsons ([email protected]) Jennifer McDaniel ([email protected]) Anne Bergstrom Lucas ([email protected]) Jean Lozach ([email protected]) Timothy G Myers ([email protected]) Qin Su ([email protected]) Sarah M Jacobs-Helber ([email protected]) Marc Salit ([email protected]) bioRxiv preprint doi: https://doi.org/10.1101/034868; this version posted January 7, 2016. -

Single-Molecule Studies of Mechanical and Helicase-Catalyzed Disruption of Nucleic Acid Duplexes
© 2017 Kevin D. Whitley SINGLE-MOLECULE STUDIES OF MECHANICAL AND HELICASE-CATALYZED DISRUPTION OF NUCLEIC ACID DUPLEXES BY KEVIN D. WHITLEY DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biophysics and Computational Biology in the Graduate College of the University of Illinois at Urbana-Champaign, 2017 Urbana, Illinois Doctoral Committee: Associate Professor Yann R. Chemla, Chair Professor Oleksii Aksimentiev Professor Paul R. Selvin Professor Taekjip Ha, Johns Hopkins University Abstract Nucleic acids (e.g. DNA, RNA) are subjected to numerous twisting, bending, and stretching forces within cells, and enzymes process them in a variety of ways. The behavior of nucleic acids in response to applied forces and enzymatic activity is therefore necessary for a fundamental understanding of biology. Furthermore, a detailed knowledge of nucleic acids and the enzymes that process them has fueled advances in bio- and nano-technology. In this thesis, we focus on two main systems: the elastic behavior of ultrashort nucleic acids and the activity of E. coli UvrD helicase. First, we use a hybrid instrument combining high-resolution optical tweezers with single-fluorophore sensitivity to observe the hybridization of ultrashort (<15 nt) DNA and RNA oligonucleotides under tension, one molecule at a time. We quantify the effect of tension on the rates of hybridization, and in doing so determine the elastic behavior of the transition state for the reaction. We then investigate the elasticity of the ultrashort oligonucleotides by observing the change in extension that takes place during hybridization. Our results enable us to produce a model describing the shear-induced fraying of base-pairs in a nucleic acid duplex. -

Enzyme-Linked Oligonucleotide Hybridization Assay for Direct Oligo Measurement in Blood Mary Y
Biology Methods and Protocols, 2019, 1–8 doi: 10.1093/biomethods/bpy014 Methods Manuscript METHODS MANUSCRIPT Enzyme-linked oligonucleotide hybridization assay for direct oligo measurement in blood Mary Y. Lorenson, Kuan-Hui Ethan Chen, and Ameae M. Walker* Division of Biomedical Sciences, School of Medicine, University of California, Riverside, CA 92521, USA *Correspondence address: Division of Biomedical Sciences, School of Medicine, University of California, Riverside, CA 92521, USA. E-mail: [email protected] Abstract Small oligonucleotides (oligos) are increasingly being utilized as diagnostics or treatments for disease. An impediment to broader use is the ability to readily measure oligos in biological fluids. Here, we describe a very straightforward assay with detection in the sub-picomole range that does not require extraction from serum/plasma or polymerization chain reaction amplification. As a result, there are no losses or errors due to sample handling, and the assay can be used to measure oligos modified in a variety of ways that increase therapeutic efficacy. The enzyme-linked oligonucleotide hybridization assay (ELOHA) is based on competition with a detection oligo for hybridization to a capture oligo covalently linked to a solid substrate. The versatility of ELOHAs is demonstrated by application to the measurement of three oligos, including two morpholino-oligos with 30-octaguanidine derivatization for efficient cell uptake. The third oligo is unmodified and has a DNA sequence equivalent to miR93. The assays have sensitivity as low as 0.28 pmol/sample reaction at 50% hybridization. Adding to clinical utility is the need for only a simple 96-well absorbance plate reader and the finding that neither EDTA nor heparin interferes with detection. -

Module 3- Lecture 1 Methods of Nucleic Acid Detection
NPTEL – Biotechnology – Genetic Engineering & Applications MODULE 3- LECTURE 1 METHODS OF NUCLEIC ACID DETECTION 3-1.1 Introduction: Nucleic acid molecules like deoxyribonucleic acids (DNA), ribonucleic acids (RNA) are basic, essential and primary molecules for all molecular biology related research. Before discussing the common methods to detect nucleic acids let us we learn about the isolation of pure form of nucleic acid. 3-1.2 Preparation Step: First important step is to establish if a given sample contains DNA or RNA and whether it is in pure form, since many samples will contain both species as well as other contaminants such as cellular proteins. Irrespective of the method applied for detection, the optimal nucleic acid isolation protocol must provide a) Reproducible result b) No degradation of the sample and c) Safer handling. In deciding what method of nucleic acid measurement is appropriate, three issues are critical: specificity, sensitivity, and interfering substances. Based on these features different detection techniques are to be followed. 3-1.3 Traditional Detection Methods: 3-1.3.1 UV spectroscopy Majority of bio-molecules intrinsically absorb light in the ultraviolet and not in the visible range. This property of UV absorbance can be used to quickly estimate the concentration and purity of DNA and RNA (also proteins) in a analytical sample. The amount of DNA in a sample can be estimated by looking at its absorbance at a wavelength of 260nm or 280nm (in the UV region). Purines and pyrimidines have absorbance maxima slightly below and above 260 respectively. Thus the absorbance maxima of different fragments of DNA vary somewhat depending on their subunit Joint initiative of IITs and IISc – Funded by MHRD Page 1 of 86 NPTEL – Biotechnology – Genetic Engineering & Applications composition. -

Using Pna Probes for Hybridization-Based Analysis of Mirnas in Capillary Electrophoresis
USING PNA PROBES FOR HYBRIDIZATION-BASED ANALYSIS OF MIRNAS IN CAPILLARY ELECTROPHORESIS MANSI ANAND A THESIS SUBMITTED TO THE FACULTY OF GRADUATE STUDIES IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF CHEMISTRY GRADUATE PROGRAM IN CHEMISTRY YORK UNIVERSITY TORONTO, ONTARIO April 2019 © Mansi Anand, 2019 i ABSTRACT MicroRNAs (miRNAs) are short non-coding RNA molecules (18‒25 nucleotides) well- known for their post-transcriptional gene regulatory roles. The pathophysiology of various dis- eases (such as neurodegenerative, cardiovascular, etc.) and in particular cancer, have been char- acterised by the abnormal cellular expression levels of miRNAs. Therefore, validation and clini- cal implementation of reliable miRNA-based biomarkers would be appreciably helpful. To uti- lize miRNA potential in disease diagnosis and prognosis, accurate and robust quantitative ap- proaches are required to analyze the sets of deregulated miRNAs (particular to a certain disease) in minimally-processed clinical samples. One such technique, developed by our lab, is direct quantitative analysis of multiple miRNA (DQAMmiR) using commercial capillary electrophore- sis (CE) setting. The hybridization-based method is direct; does not require any miRNA pro- cessing or extraction, and just uses fluorescently labeled single stranded (ss) DNA probes and two kinds of mobility shifters: (i) single-stranded binding protein (SSB) in CE run buffer (binds to the excess probes and facilitate their separation from the hybrids), (ii) different length peptide drag tags conjugated to the ssDNA probes (render different charge: size ratios amongst the hy- brids and enable their separation from each other); to simultaneously detect, separate and quanti- fy multiple miRNAs. In my study, we introduced and developed a second-generation of DQAMmiR, which is simpler than the first-generation, as it omits the addition of SSB in CE run buffer by the use of uncharged Peptide Nucleic Acid (PNA) instead of ssDNA as the hybridiza- tion probes.