Inter Simple Sequence Repeat Analysis of Genetic Diversity of Five Cultivated Pepper Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Native Peppers from Around the Globe
International Gardener Native Peppers From Around the Globe By Pat Dickey & Ray Novitske, Fairfax Master Gardeners Question: Where do peppers in Indian curries, Thai noodles, Mexican enchilladas, and Chinese . .stir-fry come from? Answer: South America Even though we associate different sizes, colors, shapes, and tastes of peppers with different nationalities and cuisines, they all originate in South America. Upon being exposed to the capsicum plant in the Caribbean, Columbus thought he had come across the peppercorn, and thus called it pepper. Peppers were spread around the world as a spice, mostly by 16th century Portuguese traders interested in the lucrative spice trade. After some time and regional cultivation, new varieties popped up and became integrated into the regional cooking we are familiar with today. This is the first in a new series which will expose us to new vegetables and ornamentals that may be growing in our international gardeners’ gardens. Aji Amarillo Native South American peppers, Aji Amarillo peppers (Capsicum baccatum) are popular in Peruvian cooking. They can be used fresh for soups and sauces, made into a chili paste, or dried. The smaller Aji ‘Chinchi’ Amarillo pepper seeds were recently introduced in the US and is a cultivar of the larger Aji Amarillo species. ‘Chinchi’ bears fruit much sooner in the season than the larger Amarillo species. The peppers are also considered rare and measure only 3 inches by one-half inch. Aji ‘Chinchi’ Amarillo peppers are fruity and full of flavor with medium- Exchange Exchange high heat. The peppers develop from green to a golden photo: ISouthern Exposure Seed www.SouthernExposure.com orange-yellow before harvest in 52 days. -

Wild Capsicum in the Area of the Amboró National Park in Bolivia
Wild Capsicum in the area of the Amboró National Park in Bolivia Claudio Dal Zovo1, Leonardo Bruno2 1 Associazione Pepperfriends, Verona, Italy 2 Associazione Pepperfriends, Roma, Italy Abstract Bolivia is believed to be the source of the genus Capsicum; possibly Capsicum chacoense Hunz. is the species closer to the ancestor of all Capsicum species. About ten species of wild Capsicum grow in Bolivia: Capsicum baccatum L. var. baccatum, Capsicum caballeroi Nee, Capsicum cardenasii Heiser & Smith, Capsicum ceratocalyx Nee, Capsicum chacoense Hunz., Capsicum coccineum (Rusby) Hunz., Capsicum eshbaughii Barboza, Capsicum eximium Hunz., Capsicum minutiflorum (Rusby) Hunz. A couple of possible new species are under investigations. Many cultivated species are also grown and sometimes present in wild forms, especially Capsicum pubescens Ruiz & Pav., Capsicum frutescens L., Capsicum baccatum L. var. pendulum (Willd.) Eshbaugh. These species are preserved in herbaria and described in articles through drawings, but few or no images are available. We wished to produce a better documentation of live plants and their details; so we planned a trip to Bolivia starting in the area where most of the less known species are concentrated. We visited the area around the Amboró National Park, from Santa Cruz de la Sierra up to Samaipata, Mairana and Comarapa (South side of the Park) and the area near Buena Vista (North side of the Park). We found populations of C.minutiflorum (Rusby) Hunz., C.caballeroi Nee, C.eximium Hunz., C.baccatum L. var. baccatum, C.coccineum (Rusby) Hunz., fully described and documented them with many detailed images. These species are well differentiated and each of them has particular characteristics. -

Reimer Seeds Catalog
LCTRONICLCTRONIC CATALOGCATALOG Drying Hot Peppers HP320‐20 ‐ Achar Hot Peppers HP321‐10 ‐ Aci Sivri Hot Peppers 85 days. Capsicum annuum. Open 85 days. Capsicum annuum. Open Pollinated. The plant produces good yields Pollinated. The plant produces good yields of 3 ¼" long by 1" wide hot peppers. Peppers of 7 ½" long by ½" wide Cayenne type hot are hot, have medium thin flesh, and turn peppers. Peppers are medium hot, have from green to deep red when mature. The medium thin flesh, and turn from light plant has green stems, green leaves, and yellowish‐green to red when mature. The white flowers. Excellent for pickling and plant has green stems, green leaves, and seasoning spice. A variety from India. United white flowers. Excellent drying, pickling, and States Department of Agriculture, PI 640826. seasoning powder. An heirloom variety from Scoville Heat Units: 27,267. Turkey. HP21‐10 ‐ Afghan Hot Peppers HP358‐10 ‐ African Fish Hot Peppers 85 days. Capsicum annuum. Open 85 days. Capsicum annuum. Open Pollinated. The plant produces good yields Pollinated. The plant produces good yields of 3" long by ½" wide Cayenne hot peppers. of 1 ½" long by ½" wide hot peppers. Peppers are very hot, have medium thin Peppers are medium‐hot, have medium thin flesh, and turn from green to red when flesh, and turn from cream white with green mature. The plant has green stems, green stripes, to orange with brown stripes, then leaves, and white flowers. Excellent for to red when mature. The plant has Oriental cuisine and for making hot pepper variegated leaves. An African‐American flakes and seasoning spice powder. -

“Caratterizzazione Di Alcune Cultivar Autoctone Peruviane Di Capsicum Sp
UNIVERSITA’ DEGLI STUDI DI PISA Facoltà di Agraria Corso di laurea specialistica in Produzioni Agroalimentari e Gestione degli Agroecosistemi Curriculum: Produzioni Agroalimentari Titolo della Tesi: “Caratterizzazione di alcune cultivar autoctone peruviane di Capsicum sp. piccante” Candidato: Relatore: Tommaso Maggiorelli Prof. Lorenzo Guglielminetti Anno Accademico 2012/2013 Indice Ringraziamenti pag.1 Scopo della tesi pag.2 Parte I – Introduzione pag.3 1.1 Cenni storici sul peperoncino 1.2 Il peperoncino, prima e dopo l’avanzata europea 1.3 Le specie di Capsicum 1.4 Il Capsicum 1.4.1 Capsicum annuum var. annuum L. 1.4.2 Capsicum chinense Jacq. 1.4.3 Capsicum frutescens L. 1.4.4 Capsicum baccatum var. pendulum Esh. 1.4.5 Capsicum pubescens Ruiz & Pavon 1.5 Origine ed evoluzione del Capsicum 1.6 Caratteri botanici e varietali di C. annuum, C.frutescens e C. chinense 1.7 Importanza industriale del peperoncino 1.8 Sapore piccante 1.9 Proprietà Parte II – Materiali e metodi pag.29 2.1 Coltivazione e tecniche agronomiche 2.2 Estrazione e determinazione capsaicina 2.2.1 La scala di Scoville 2.2.2 Estrazione 2.2.3 Panel Test Parte III – Risultati e conclusione pag.39 3.1 Peperoncini peruviani I. Aji Amarillo II. Aji Limo Rosso III. Aji Red IV. Aji Rojo V. Aji Unas de Galina VI. Rocoto Manzano VII. Capezzolo di Scimmia VIII. Charapita 3.2 Conclusioni pag. 76 Bibliografia pag. 78 Ringraziamenti Desidero innanzitutto ringraziare il Professor Amedeo Alpi e il Professor Lorenzo Guglielminetti per il supporto fornito e per il tempo dedicato al perfezionamento della tesi. -

Hot Peppers and Specialty Sweet Peppers
Center for Crop Diversification Crop Profile CCD-CP-101 Hot Peppers and Specialty Sweet Peppers Cheryl Kaiser1 and Matt Ernst2 Introduction Hot peppers, also known as chili (or chile) peppers, owe most of their “heat” or pungency to a chemical substance called capsaicin. This chemical is concen- trated in the cross walls of the fruit and around the developing seeds. Chili peppers can be mild to fiery hot, depending on the amount of capsaicin present. The amount of capsaicin in peppers is measured in Scoville Heat Units (SHU). Currently, the hottest pep- per is considered to be the ‘Carolina Reaper’ which has 2.2 million SHUs. A combination of genetics and environment are responsible for the amount of heat in hot peppers. Peppers that do not contain capsa- icin, such as bell peppers (0 SHUs), are considered “sweet.” In addition to the hot types, other specialty HABANERO PEPPERS peppers include sweet varieties of unusual shape, size HABANEROS (Capsicum chinense) are extremely hot and/or color. peppers that are small and lantern-shaped. They are light green to bright orange when ripe. Types of Hot and Specialty Sweet Peppers Unless otherwise noted, the following peppers are ITALIAN or CUBANELLE types are sweet to mildly hot, classified as the speciesCapsicum annuum. long, and somewhat flattened. These flavorful peppers change from yellow-green to orange, and then to red ANAHEIM peppers, also known as NEW MEXICAN CHILE, as they ripen. are a mild to hot pepper that are considerably longer than jalapeños. They are bright green to red when JALAPEÑO peppers range from sweet to mild to very fresh and brownish red when dried. -

Capsicum'' 1 Capsicum
''Capsicum'' 1 Capsicum Capsicum Fruit and longitudinal section Scientific classification Kingdom: Plantae (unranked): Angiosperms (unranked): Eudicots (unranked): Asterids Order: Solanales Family: Solanaceae Subfamily: Solanoideae Tribe: Capsiceae Genus: Capsicum [1] L. Species [2] See text For the fruit, see: Chili pepper For the heat simulating chemical in Chili pepper, see: Capsaicin Capsicum is a genus of flowering plants in the nightshade family, Solanaceae. Its species are native to the Americas, where they have been cultivated for thousands of years by the people of the tropical Americas, and are now cultivated worldwide. Some of the members of Capsicum are used as spices, vegetables, and medicines. The fruit of Capsicum plants have a variety of names depending on place and type. They are commonly called chilli pepper, red or green pepper, or sweet pepper in Britain, and typically just capsicum in Australian, New Zealand, and Indian English. The large mild form is called bell pepper in the U.S. and Canada. They are called paprika in some other countries (although paprika can also refer to the powdered spice made from various capsicum fruit). The generic name is derived from the Greek word καπτο (kapto), meaning "to bite" or "to swallow."[3] The original Mexican term, chilli (now chile in Mexico) came from the Nahuatl word chilli or xilli, referring to a larger Capsicum variety cultivated at least since 3000 BC, as evidenced by remains found in pottery from Puebla and Oaxaca.[4] ''Capsicum'' 2 Capsaicin in capsicum The fruit of most species of Capsicum contains capsaicin (methyl vanillyl nonenamide), a lipophilic chemical that can produce a strong burning sensation in the mouth of the unaccustomed eater. -

The Aloe, Succulent and Cactus Society Outing Circular
ORTHOLOPHA The Newsletter of the Aloe, Cactus and Succulent Society of Zimbabwe PO Box CY300, Causeway [email protected] Issue 13-1 January 2013 INTO THE NEW YEAR Your Committee Very many thanks to John and Pat for a delightful Chairman - Manfred Spindler – 0712 416 722 Christmas function. We all appreciated the effort you two made to make the day such a success, and Vice-chairman - Malcolm Thackray - it was a pleasure to observe the changes in 303494 Secretary/treasurer - Lynn & Bill Kinsey – landscaping that have taken place. And, John, we 0772 782 493, 0776 404 950 or 302812 look forward to seeing what that amazing new greenhouse is going to look like when it is full. Committee members: Val & Hamish Brown - 0772 423 344 or After very much below-average rainfall in November 883724 and December, January is trying to make up for it. Ros Houghton - 0772 115 364 or 333975 By mid-month, we had already had 90% of our Rob & Sheila Jarvis – 0779 888 180 or 0772 262 average January rain and it just doesn’t seem to 150 want to stop. The boreholes are singing. So, if you Mike Kimberley – 339175 can get out without sinking, there is still plenty of Hans Wolbert – 581364, 573592 or 0772 good gardening weather ahead. See below for the 653 110 available offerings from the seed bank. Sunday, 27th January 2013 The first outing of the New Year will be at the garden of the Hooks—Sharon and Ed—in Borrowdale. The address: 34 Kingsmead Road West, Borrowdale Phones: 0772 240 442 or 870877 The time: 10.00 Details are below. -

Hot Pepper (Capsicum Spp.) – Important Crop on Guam
Food Plant Production June 2017 FPP-05 Hot Pepper (Capsicum spp.) – Important Crop on Guam Joe Tuquero, R. Gerard Chargualaf and Mari Marutani, Cooperative Extension & Outreach College of Natural & Applied Sciences, University of Guam Most Capsicum peppers are known for their spicy heat. Some varieties have little to no spice such as paprika, banana peppers, and bell peppers. The spice heat of Capsicum peppers are measured and reported as Scoville Heat Units (SHU). In 1912, American pharmacist, Wilbur Scoville, developed a test known as the, Scoville Organoleptic Test, which was used to measure pungency (spice heat) of Capsicum peppers. Since the 1980s, pungency has been more accurately measured by high-performance liquid chromatography Source: https://phys.org/news/2009-06-domestication- (HPLC). HPLC tests result in American Spice Trade capsicum-annuum-chile-pepper.html Association (ASTA) pungency units. ASTA pungency Introduction units can be converted to SHU. Table 2 displays Sco- Hot pepper, also known as chili, chilli, or chile pepper, ville Heat Units of various popular Capsicum peppers is a widely cultivated vegetable crop that originates (Wikipedia, 2017). from Central and South America. Hot peppers belong to the genus Capsicum. There are over 20 species under the genus Capsicum. There are five major domesticated species of peppers that are commercially cultivated (Table 1), and there are more than 50,000 varieties. Fig. 1 depicts a unqiue, citrus-flavored variety of Capsicum baccatum hot pepper, known as Lemon Drop (aji-type), popular for seasoning in Peru (Wikipedia, 2017). Table 1. The five major domesticated Capsicum species of pepper with examples of commonly known types of pepper. -

2018 Mole Pepper Variety Trial
Midwest Vegetable Trial Report for 2018 2018 Mole Pepper Variety Trial Ben Phillips, Michigan State University Extension One Tuscola St, Suite 100A, Saginaw, MI 48607 Office: 989.758.2502 Email: [email protected] This project was undertaken with a client who wanted to make mole (pronounced “moh- lay”) sauces from Michigan-grown and Michigan-dried poblano (dried ancho), chilaca (dried pasilla), mirasol (dried guajillo) peppers. The peppers must be fully ripened before drying for the right flavor. Therefore, the main interest of this study was to determine which varieties would yield the most ripe colored fruit before the first frost. A secondary objective of this project was to dry the peppers, covered in a separate report. Materials and Methods The mole pepper variety trial was planted at the Saginaw Valley Research and Extension Center (43.399097, -83.694497, Frankenmuth, Michigan). The soil type was a Tappan-Londo loam with a poor-moderate drainage class. On 30 May 180 pounds 46-0-0 was preplant incorporated, resulting in ~80 lb N per acre. The same day, 13 varieties were transplanted in a completely randomized block design with four replications. Sakata (SK), PanAmerican (PAN), Siegers and Harris seed companies and private Wisconsin breeder, James Nienhuis (JN), donated seeds to the trial. Varieties donated by Siegers are owned by Seminis (SG) and US Agriseed (UA). The variety donated by Harris is owned by Seminis. Transplants were started by a local greenhouse on 7 April in 72-cell trays and were eight inches tall at transplanting. Plots consisted of a single row 20 ft long. -
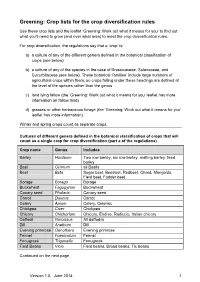
Greening: Crop Lists for the Crop Diversification Rules
Greening: Crop lists for the crop diversification rules Use these crop lists and the leaflet ‘Greening: Work out what it means for you’ to find out what you’ll need to grow (and over what area) to meet the crop diversification rules. For crop diversification, the regulations say that a ‘crop’ is: a) a culture of any of the different genera defined in the botanical classification of crops (see below) b) a culture of any of the species in the case of Brassicaceae, Solanaceae, and Cucurbitaceae (see below). These botanical ‘families’ include large numbers of agricultural crops within them, so crops falling under these headings are defined at the level of the species rather than the genus. c) land lying fallow (the ‘Greening: Work out what it means for you’ leaflet has more information on fallow land) d) grasses or other herbaceous forage (the ‘Greening: Work out what it means for you’ leaflet has more information) Winter and spring crops count as separate crops. Cultures of different genera defined in the botanical classification of crops that will count as a single crop for crop diversification (part a of the regulations) Crop name Genus Includes Barley Hordeum Two row barley, six row barley, malting barley, feed barley Basil Ocimum all Basils Beet Beta Sugar beet, Beetroot, Redbeet, Chard, Mangolds, Field beet, Fodder beet Borage Borago Borage Buckwheat Fagopyrum Buckwheat Canary seed Phalaris Canary seed Carrot Daucus Carrot Celery Apium Celery, Celeriac Chickpea Cicer Chickpea Chicory Chichorium Chicory, Endive, Radiccio, Italian chicory Daffodil Narcissus All daffodils Dill Anethum Dill Evening primrose Oenothera Evening primrose Fennel Foeniculum Fennel Fenugreek Trigonella Fenugreek Field Beans Vicia Field beans, Broad beans, Tic beans Continued on the next page. -

Capsaicin, Phenols and Flavonoids Quantification in Leskovac and Habanero Hot Peppers
Advanced technologies 9(1) (2020) 21-26 CAPSAICIN, PHENOLS AND FLAVONOIDS QUANTIFICATION IN LESKOVAC AND HABANERO HOT PEPPERS Dragan T. Veličković1,*, Mirjana N. Virijević1, Ljiljana P. Stanojević2, Jelena S. Stanojević2, Ivana S. Mošić3, Slađana S. Golubović1, Sanja R. Perić1, 4 Grozdan R. Stamenković (ORIGINAL SCIENTIFIC PAPER) UDC 633.842:547.9:66.061 1Academy of Professional Studies South Serbia, Department of Agricultural and Food Studies, Prokuplje, Serbia 2Faculty of Technology, University of Niš, Leskovac, Serbia 3Aromatika doo, Niš, Serbia 4"Zdravlje-Actavis" Company, Leskovac, Serbia Both as food and spice, pepper is unavoidable on the tables in Serbia and many other countries. Because of its popularity, the contents of capsaicin, total phenols and total flavonoids, as well as the antioxidant activity of hot peppers extracts (Leskovac hot pepper, so called “džinka” and chili Habanero pepper) were examined in this paper. Keywords: Capsicum annuum L., Cap- The extracts were obtained by Soxhlet extraction with 96% v/v ethanol. The content sicum chinense Jacq., Capsaicin, Extrac- of capsaicin in dry extracts of the studied hot peppers was calculated by the HPLC tion method (13.038 mg/g d.e. for “džinka” and 76.516 mg/g d.e. for Habanero pepper, respectively). Total phenol and total flavonoids contents were examined by the UV- VIS method. Leskovac “džinka” was characterized by both higher contents (47.17 mgGAE/g d.e. and 14.64 mgGAE/g d.e., respectively) in comparison to chili Haba- nero pepper (29.43 mgGAE/g d.e. and 0.25 mgGAE/g d.e., respectively). DPPH test was used for the antioxidant activity determination. -

Characterization of Capsicum Species Using Anatomical and Molecular Data
Characterization of Capsicum species using anatomical and molecular data G.B. Dias1, V.M. Gomes2, T.M.S. Moraes1, U.P. Zottich2, G.R. Rabelo1, A.O. Carvalho2, M. Moulin3, L.S.A. Gonçalves3, R. Rodrigues3 and M. Da Cunha1 1Setor de Biologia Vegetal, Laboratório de Biologia Celular e Tecidual, Centro de Biociências e Biotecnologia, Universidade Estadual do Norte Fluminense, Rio de Janeiro, RJ, Brasil 2Laboratório de Fisiologia e Bioquímica de Microrganismos, Universidade Estadual do Norte Fluminense, Rio de Janeiro, RJ, Brasil 3Laboratório de Melhoramento Genético Vegetal, Universidade Estadual do Norte Fluminense, Rio de Janeiro, RJ, Brasil Corresponding author: M. Da Cunha E-mail: [email protected] Genet. Mol. Res. 12 (4): 6488-6501 (2013) Received August 7, 2012 Accepted November 22, 2012 Published February 28, 2012 DOI http://dx.doi.org/10.4238/2013.February.28.29 ABSTRACT. Capsicum species are frequently described in terms of genetic divergence, considering morphological, agronomic, and molecular databases. However, descriptions of genetic differences based on anatomical characters are rare. We examined the anatomy and the micromorphology of vegetative and reproductive organs of several Capsicum species. Four Capsicum accessions representing the species C. annuum var. annuum, C. baccatum var. pendulum, C. chinense, and C. frutescens were cultivated in a greenhouse; leaves, fruits and seeds were sampled and their organ structure analyzed by light and scanning electronic microscopy. Molecular accession characterization was made using ISSR markers. Polymorphism was observed among tector trichomes and also in fruit color and shape. High variability among accessions was detected by ISSR markers. Although the species studied Genetics and Molecular Research 12 (4): 6488-6501 (2013) ©FUNPEC-RP www.funpecrp.com.br Characterization of Capsicum species 6489 present a wide morphological and molecular variability, this variability was not reflected in anatomical features.