On-Site Detection of Xylella Fastidiosain Host Plants and in “Spy Insects”
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nerium Oleander Linn. (Kaner)
Available online on www.ijppr.com International Journal of Pharmacognosy and Phytochemical Research 2014; 6(3): 593-597 ISSN: 0975-4873 Review Article A Review on: Nerium oleander Linn. (Kaner) *Chaudhary Kiran1, Prasad D.N.2 1K.C. Institute of Pharmaceutical Sciences, Pandoga, Distt.Una (H.P.) 2Shivalik College of Pharmacy, Naya Nangal, Distt.Ropar, Punjab Available Online: 1st September 2014 ABSTRACT Nerium oleander is an evergreen shrub or small tree in the dogbane family Apocyanaceae. It is commonly known as oleander but has many other names like Nerium indicum mill. and Nerium odorum soland. It bears flowers in clusters with white, pink, yellow and red colours. It contains plumericin, alpha-amyrin, beta-sitosterol, kaempferol, cardioactive glycosides named Odorosides A-H obtained from the root bark. Leaves contain the cardiac glycosides kaneroside, neriumoside, digitoxigenin, alpha –L-olendroside -5α-adynerin and other glycosides. Odorosides are cardioactive glycosides. Gentiobiosyl –oleandrin, Odoroside A and Oleandrin were the main glycosides identified. It has potent cardiotonic activity, digitalis like effect on heart. It has been reported to have effective against skin diseases,wound infections, cancer, diabetes, inflammation and CNS depression. All parts of the plant are poisonous in nature which can be treated by the use of activated charcoal.Topical preparation containing Nerium extract can be used as antiageing cream. Keywords : Nerium oleander, Nerium indicum, Oleander, Cardiotonic, Odorosides , Antibacterial, Antiageing INTRODUCTION Nepal westwards to Kashmir upto 1950m, extending to Taxonomic classification Baluchistan, Afghanistan and found throughout India in Phyllum- Plantae gardens .The white and red flowered variety is equated Class/ Subphyllum- Angiosperms with Nerium indicum. -

A Dunedin Garden
VIREYA VINE ISSUE #82, OCTOBER 2007 PUBLISHED BY THE EDUCATION COMMITTEE OF THE RHODODENDRON SPECIES FOUNDATION R.S.F. PO BOX 3798, FEDERAL WAY, WA. 98063 E. White Smith, Editor From Daphne and Gavin Clark Dunedin, New Zealand Published with permission from the Dunedin Bulletin Feb. 2007 RHODODENDRON lowii (Subsection Vireya) More information about R. lowii relting to the piece in VV81 Addendum – February 2007 Following on from the previous article the long cane produced four shoots. Two were given to a very keen propagator to strike and we tried to strike two, but they all failed. However a good quantity of seed was collected at the end of January, 2005 and again this was shared. Our seed was kept in cool storage and eventually sown in September 2005. A small transparent plastic container with a clear lid was used with the seed sprinkled on to damp, sterilized sphagnum moss. The seeds germinated very well and some of the seedlings were transferred into compost but despite tender care died. The remaining seedlings stayed in the plastic container for some considerable time until recently when they were very carefully removed to individual peat pots into a specially mixed compost. Nine pots are housed in a clear, plastic lidded container 340mm x 300mm which stands on a wide kitchen windowsill with excellent light but no direct sunlight and hopefully some of these will survive. In the meantime the parent plant is flourishing, the two basal shoots are now 800mm high with the original cane cut down to 750mm. It is still in its container and now occupies a choice place among other vireya species in a wooded area beneath 50 year old camellias and rhododendrons which have been pruned to provide an excellent canopy, with morning sun, filtered afternoon sunlight, together with a degree of humidity. -

European Academic Research
EUROPEAN ACADEMIC RESEARCH Vol. IV, Issue 10/ January 2017 Impact Factor: 3.4546 (UIF) ISSN 2286-4822 DRJI Value: 5.9 (B+) www.euacademic.org Evidences from morphological investigations supporting APGIII and APGIV Classification of the family Apocynaceae Juss., nom. cons IKRAM MADANI Department of Botany, Faculty of Science University of Khartoum, Sudan LAYALY IBRAHIM ALI Faculty of Science, University Shandi EL BUSHRA EL SHEIKH EL NUR Department of Botany, Faculty of Science University of Khartoum, Sudan Abstract: Apocynaceae have traditionally been divided into into two subfamilies, the Plumerioideae and the Apocynoideae. Recently, based on molecular data, classification of Apocynaceae has undergone considerable revisions. According to the Angiosperm Phylogeny Group III (APGIII, 2009), and the update of the Angiosperm Phylogeny Group APG (APGIV, 2016) the family Asclepiadaceae is now included in the Apocynaceae. The family, as currently recognized, includes some 1500 species divided in about 424 genera and five subfamilies: Apocynoideae, Rauvolfioideae, Asclepiadoideae, Periplocoideae, and Secamonoideae. In this research selected species from the previous families Asclepiadaceae and Apocynaceae were morphologically investigated in an attempt to distinguish morphological important characters supporting their new molecular classification. 40 morphological characters were treated as variables and analyzed for cluster of average linkage between groups using the statistical package SPSS 16.0. Resulting dendrograms confirm the relationships between species from the previous families on the basis of their flowers, fruits, 8259 Ikram Madani, Layaly Ibrahim Ali, El Bushra El Sheikh El Nur- Evidences from morphological investigations supporting APGIII and APGIV. Classification of the family Apocynaceae Juss., nom. cons and seeds morphology. Close relationships were reported between species from the same subfamilies. -

A Hydroalcoholic Extract from the Leaves of Nerium Oleander Inhibits Glycolysis and Induces Selective Killing of Lung Cancer Cells
Original Papers A Hydroalcoholic Extract from the Leaves of Nerium oleander Inhibits Glycolysis and Induces Selective Killing of Lung Cancer Cells Authors José Manuel Calderón-Montaño1, Estefanía Burgos-Morón1, Manuel Luis Orta2, Santiago Mateos2, Miguel López-Lázaro1 Affiliations 1 Department of Pharmacology, Faculty of Pharmacy, University of Seville, Seville, Spain 2 Department of Cell Biology, Faculty of Biology, University of Seville, Seville, Spain Key words Abstract consumption and lactate production) in A549 l" Nerium oleander ! cells, comparable to that of the glycolysis inhibitor l" Apocynaceae Recent evidence suggests that cardiac glycosides dichloroacetate (currently in clinical develop- l" cardiac glycosides might be used for the treatment of cancer. The or- ment for cancer therapy). Because platinum com- l" cardiotonic steroids namental shrub Nerium oleander has been used in pounds are widely used in the treatment of lung l" anticancer traditional medicine for treating several disorders cancer, we tested the cytotoxicity of several com- including cancer, and extracts from the leaves of binations of cisplatin with the extract and found a this plant have already entered phase I clinical tri- moderate synergism when Nerium oleander ex- als. In this communication, we have prepared a tract was administered after cisplatin but a mod- hydroalcoholic extract from the leaves of Nerium erate antagonism when it was added before cis- oleander (containing 4.75 ± 0.32% of cardenolides) platin. Our results suggest that extracts from Ne- and have assessed its cytotoxic activity in A549 rium oleander might induce anticancer effects in lung cancer cells vs. MRC5 nonmalignant lung fi- patients with lung cancer and support their possi- broblasts. -

D.1A Floral and Faunal Compendia
Appendix D.1a Floral and Faunal Compendia APPENDIX D.1a FLORAL AND FAUNAL COMPENDIA INTRODUCTION TO FLORAL AND FAUNAL SURVEY Expected site use by wildlife is derived from survey information combined with documented habitat preferences of regional wildlife species, which, whether or not recorded during the survey, are considered likely to include the project area within their range. Habitat designations used in this report are according to the classification system of Holland (1986). Floral taxonomy used in this report follows the Jepson Manual (Hickman 1993), with updates in accordance to the online Jepson Interchange where known. Common plant names, where not available from Munz (1974), are taken from Abrams (1923), Robbins, et al. (1951), Collins (1972), Niehaus and Ripper (1976) and Muns (1983). Vertebrates identified in the field by sight, calls, tracks, scat or other signs are cited according to the nomenclature of Jennings (1983) for amphibians and reptiles; AOU (1983) for birds; and Jones, et al. (1982) for mammals. Butterflies observed or collected in the field were identified with the help of Garth and Tilden (1986) and Tilden and Smith (1986). FLORAL COMPENDIUM1 LEGEND * Nonnative @ Ornamental/Landscape VASCULAR PLANTS CONIFERAE PINACEAE - PINE FAMILY * Pinus halepensis Aleppo Pine ANGIOSPERMAE (DICOTYLEDONS) ANACARDIACEAE - SUMAC FAMILY Malosma laurina laurel sumac Rhus ovata sugar bush * Schinus molle Peruvian pepper-tree Toxicodendron diversilobum poison-oak APOCYNACEAE - DOGBANE FAMILY * Vinca major periwinkle * Nerium oleander oleander ASTERACEAE - SUNFLOWER FAMILY Baccharis pilularis coyote brush Baccharis salicifolia mulefat Helianthus gracilentus slender sunflower * Picris echioides bristly ox-tongue * Silybum marianum milk thistle * Sonchus asper prickly sow-thistle * Sonchus oleraceus common sow-thistle Stephanomeria virgata twiggy wreathplant BRASSICACEAE - MUSTARD FAMILY * Brassica nigra black mustard * Sisymbrium officinale hedge-mustard CAPRIFOLIACEAE - HONEYSUCKLE FAMILY Sambuccus. -

Calaveras Nursery Inc. (925)862-2286 1000 Calaveras Rd
Calaveras Nursery Inc. (925)862-2286 1000 Calaveras Rd. Sunol, CA 94586 Plant availability. "X" denotes "in stock" or "can be ordered in". Container Size SHRUBS 1gal 5gal 15gal 24" Abelia grandiflora "White Glossy Abelia" X X Abelia grandiflora "Edward Goucher Pink Abelia" X X X Abelia Grandiflora "Francis Mason" X Abelia Grandiflora "Kaleidoscope" Abelia Grandiflora "Confetti" X Acanthus Mollis "Bears Breech" X X Achillia "Yarrow" *assorted varieties." X Agapanthus Africanus "Blue" X X Agapanthus Africanus "White" X X Agapanthus "X" Midnight Blue X Agapanthus "X" Storm Cloud X Agapanthus "Dwarf Peter Pan White" X X Agapanthus "Dwarf Peter Pan Blue" X X Ajuga *assorted varieties* X Alyogne Huegelii "Blue Hibiscus" X X Anisodontea X "Taras Pink" X X Arctostaphyllus Densiflora "Howard McMinn Manzanita" X X X Arctostaphyllus "Emerald Carpet Manzanita" X X Armeria "Sea Pink" X Artemesia "Poweys Castle" X Aucuba Japonica "Gold Spot" X X Azalea *many varieties available.* X X X Baccharis Pilularis "Twin Peaks" Dwarf Coyote Brush X Bacopa "Gullivers White" X Bamboo "Clumping Buddahs Belly" X Bamboo "Clumping Timber" X Berberis Thunbergii "Red Japanese Barberry" X X Berberis Thunbergii "Crimson Pygmy Barberry" X X Berberis Thunbergii "Rose Glow Barberry" X X Bougainvillea "Assorted colors" X Bergenia "Winter Glow" X Brunsfelsia Floribunda "Yesterday/Today/Tomorrow" X Buddleia Davidii "Black Knight" Butterfly Bush X Buxus Microphylla Japonica "Japanese Boxwood" X X Buxus Microphylla Japonica "Green Beauty Japanese Boxwood" X Buxus Microphylla Japonica "Winter Gem Boxwood" X Buxus Sempervirens "Suffruticos" True Dwarf Boxwood X X Callistemon Citrinus "Red Bottle Brush" X X X Callistemon Viminalis "Little John" Dwarf Bottlebrush X X Ceanothus G. -

Apocynaceae-Apocynoideae)
THE NERIEAE (APOCYNACEAE-APOCYNOIDEAE) A. J. M. LEEUWENBERG1 ABSTRACT The genera of tribe Nerieae of Apocynaceae are surveyed here and the relationships of the tribe within the family are evaluated. Recent monographic work in the tribe enabled the author to update taxonomie approaches since Pichon (1950) made the last survey. Original observations on the pollen morphology ofth egener a by S.Nilsson ,Swedis h Natural History Museum, Stockholm, are appended to this paper. RÉSUMÉ L'auteur étudie lesgenre s de la tribu desNeriea e desApocynacée s et évalue lesrelation s del a tribu au sein de la famille. Un travail monographique récent sur la tribu a permit à l'auteur de mettre à jour lesapproche s taxonomiques depuis la dernière étude de Pichon (1950). Lesobservation s inédites par S. Nilsson du Muséum d'Histoire Naturelle Suédois à Stockholm sur la morphologie des pollens des genres sontjointe s à cet article. The Apocynaceae have long been divided into it to generic rank and in his arrangement includ two subfamilies, Plumerioideae and Apocynoi- ed Aganosma in the Echitinae. Further, because deae (Echitoideae). Pichon (1947) added a third, of its conspicuous resemblance to Beaumontia, the Cerberioideae, a segregate of Plumerioi it may well be that Amalocalyx (Echiteae— deae—a situation which I have provisionally ac Amalocalycinae, according to Pichon) ought to cepted. These subfamilies were in turn divided be moved to the Nerieae. into tribes and subtribes. Comparative studies Pichon's system is artificial, because he used have shown that the subdivision of the Plume the shape and the indumentum of the area where rioideae is much more natural than that of the the connectives cohere with the head of the pistil Apocynoideae. -
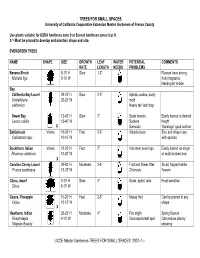
UCCE Master Gardeners TREES for SMALL SPACES 2007--1
TREES FOR SMALL SPACES University of California Cooperative Extension Master Gardeners of Fresno County Use plants suitable for USDA hardiness zone 9 or Sunset hardiness zones 8 or 9. X = Must be pruned to develop and maintain shape and size. EVERGREEN TREES NAME SHAPE SIZE GROWTH LEAF WATER POTENTIAL COMMENTS RATE LENGTH NEEDS PROBLEMS Banana Shrub 8-15’ H Slow 1-3” Flowers have strong Michelia figo 5-10’ W fruity fragrance Needs part shade Bay California Bay Laurel 20-25’ H Slow 2-5” Aphids, scales, sooty 20-25’ W mold Umbellularia californica Heavy fall leaf drop Sweet Bay 12-40’ H Slow 3” Scale insects Easily trained to desired Laurus nobilis 12-40’ W Suckers height X Sunscald ‘Saratoga” good cultivar Bottlebrush Varies 10-30’ H Fast 2-3” Attracts bees Size and shape vary Callistemon spp. 10-15’ W with species Buckthorn, Italian Varies 10-20’ H Fast 2” Volunteer seedlings Easily trained as single 10-20’ W or multi-trunked tree Rhamnus alaternus X Carolina Cherry Laurel 20-30’ H Moderate 2-4” Fruit and flower litter Small, fragrant white 15-25’ W Chlorosis flowers Prunus caroliniana Citrus, dwarf 8-10’ H Slow 3” Scale, aphid, ants Frost sensitive Citrus 8-10’ W Guava, Pineapple 15-20’ H Fast 2-3” Messy fruit Can be pruned to any Feijoa 10-15’ W shape X Hawthorn, Indian 20-25’ H Moderate 4” Fire blight Spring flowers Rhaphiolepis 8-10’ W Cercospora leaf spot Can reduce size by ‘Majestic Beauty’ shearing UCCE Master Gardeners TREES FOR SMALL SPACES 2007--1-- EVERGREEN TREES—Cont. -

(12) United States Patent (10) Patent No.: US 9,017,738 B2 Efrahimi (45) Date of Patent: Apr
US009017738B2 (12) United States Patent (10) Patent No.: US 9,017,738 B2 Efrahimi (45) Date of Patent: Apr. 28, 2015 (54) COMPOSITION FORTREATING (56) References Cited HEMORRHODS U.S. PATENT DOCUMENTS (71) Applicant: Eyal Efrahimi, Jerusalem (IL) 201 1 O274772 A1 ck 1 1, 2011 Kucukay et al. 424,727 (72) Inventor: Eyal Efrahimi, Jerusalem (IL) FOREIGN PATENT DOCUMENTS (*) Notice: Subject to any disclaimer, the term of this IT 2002-MI2263 A1 * 4, 2004 patent is extended or adjusted under 35 U.S.C. 154(b) by 0 days. * cited by examiner (21) Appl. No.: 13/769,848 Primary Examiner — Susan Hoffman (74) Attorney, Agent, or Firm — Mark M. Friedman (22) Filed: Feb. 19, 2013 (57) ABSTRACT (65) Prior Publication Data - The composition for treating hemorrhoids includes dry com US 2014/O234453 A1 Aug. 21, 2014 ponents: oleander (nerium Oleander), myrtle (myrtus commu mis)and bay laurel (laurus nobilis), which are then mixed into (51) Int. Cl. a homogenous solution with olive oil. In the preferred A6 IK 36/63 (2006.01) embodiment, the composition for treating hemorrhoids A6 IK 36/6 (2006.01) includes the following components: 30%–70% by weight of A6 IK36/54 (2006.01) olive oil, 27%-44% by weight of oleander (nerium oleander), A6 IK 36/24 (2006.01) 15%-25% percentage by weight of myrtle (myrtus communis) (52) U.S. Cl. and 5%-15% percentage by weight of bay laurel (laurus CPC ................. A61K 36/63 (2013.01); A61 K36/54 nobilis). For the most preferred embodiment, for 1 liter (about (2013.01); A61K 36/61 (2013.01); A61K 36/24 1000 cubic centimeters) of olive oil, 1000 cubic centimeters (2013.01) (cc) of the dry components of the composition may include (58) Field of Classification Search 70% percentage by weight of oleander (nerium oleander), CPC ...... -

ISSN: 2320-5407 Int. J. Adv. Res. 5(8), 486-493 RESEARCH ARTICLE
ISSN: 2320-5407 Int. J. Adv. Res. 5(8), 486-493 Journal Homepage: - www.journalijar.com Article DOI: 10.21474/IJAR01/5081 DOI URL: http://dx.doi.org/10.21474/IJAR01/5081 RESEARCH ARTICLE THEVETIA PERUVIANA: A MULTIPURPOSE MEDICINAL PLANT- A REVIEW. *Tabrez Ahmad1, Abdulhamid Tahir Hamid2, Anuradha Sharma1 and Uma Bhardwaj3. 1. Department of Biotechnology, School of Life and Applied Sciences, Baba Farid Institute of Technology, Dehradun, Uttarakhand, India. 2. School of Biotechnology, Maharaj Vinayak Global University, Jaipur, Rajasthan, India. 3. Dean Research, Swami Rama Himalayan University, Dehradun, Uttarakhand, India. …………………………………………………………………………………………………….... Manuscript Info Abstract ……………………. ……………………………………………………………… Manuscript History Plant based therapeutics play an important role in the public health care system of any nation. The plant Thevetia peruviana belongs to the Received: 05 June 2017 family Apocynaceae. The plant have a significant place in traditional Final Accepted: 07 July 2017 system of medicine of Central and South America and tropical Asia. A Published: August 2017 number of different classes of secondary metabolites are present in Key words:- Thevetia peruviana including alkaloids, flavonoids, steroids, cardiac Thevetia peruviana, Apocynaceae, glycosides, terpenoids, tannins, saponins etc. Several researchers had Cardiac glycosides, identified various pharmacological activities in different parts of the Ethanopharmacology plant viz. seeds, flowers, bark, fruits, leaves. This review covers detailed ethanopharmacology, toxicology and bioactivities of Thevetia peruviana. Copy Right, IJAR, 2017,. All rights reserved. …………………………………………………………………………………………………….... Introduction:- Our planet is home for enormous medicinal plants. Mankind use of medicinal plants is not new rather dates back to thousands of years. In fact, ancient man was very dependent on green plants for his day-to-day needs of medicaments. -

Milkweeds a Conservation Practitioner’S Guide
Milkweeds A Conservation Practitioner’s Guide Plant Ecology, Seed Production Methods, and Habitat Restoration Opportunities Brianna Borders and Eric Lee-Mäder The Xerces Society FOR INVERTEBRATE CONSERVATION The Xerces Society for Invertebrate Conservation 1 MILKWEEDS A Conservation Practitioner's Guide Brianna Borders Eric Lee-Mäder The Xerces Society for Invertebrate Conservation Oregon • California • Minnesota • Nebraska North Carolina • New Jersey • Texas www.xerces.org Protecting the Life that Sustains Us The Xerces Society for Invertebrate Conservation is a nonprofit organization that protects wildlife through the conservation of invertebrates and their habitat. Established in 1971, the Society is at the forefront of invertebrate protection, harnessing the knowledge of scientists and the enthusiasm of citizens to implement conservation programs worldwide. The Society uses advocacy, education, and applied research to promote invertebrate conservation. The Xerces Society for Invertebrate Conservation 628 NE Broadway, Suite 200, Portland, OR 97232 Tel (855) 232-6639 Fax (503) 233-6794 www.xerces.org Regional offices in California, Minnesota, Nebraska, New Jersey, North Carolina, and Texas. The Xerces Society is an equal opportunity employer and provider. © 2014 by The Xerces Society for Invertebrate Conservation Acknowledgements Funding for this report was provided by a national USDA-NRCS Conservation Innovation Grant, The Monarch Joint Venture, The Hind Foundation, SeaWorld & Busch Gardens Conservation Fund, Disney Worldwide Conservation Fund, The Elizabeth Ordway Dunn Foundation, The William H. and Mattie Wat- tis Harris Foundation, The CERES Foundation, Turner Foundation Inc., The McCune Charitable Founda- tion, and Xerces Society members. Thank you. For a full list of acknowledgements, including project partners and document reviewers, please see the Acknowledgements section on page 113. -

Aphis Nerii, One Host-Specialist Ecosystem at Intercontinental Scale
WAGENINGEN UNIVERSITY LABORATORY OF ENTOMOLOGY Asclepias syriaca – Aphis nerii, One host-specialist ecosystem at intercontinental scale C. Rogé, Msc Plant sciences Minor and Major Entomology thesis, No: 010.10 No ............................................ 85 09 06 701 060 Name ...................................................... Cyril Rogé Study program ....................................... Msc Plants Sciences Period .......................................................2010-2011 Thesis/Internship ..................... ENT 80436 + ENT 80424 (+ 6) Thesis number ............................................................. 010.10 1e Examinator ......................................................... M. Dicke 2e Examinator ............................................................R. Gols Thesis title : Asclepias syriaca – Aphis nerii, one host-specialist ecosystem at intercontinental scale Key words : Milkweed – Danaus plexippus – Induction – Enemy release hypothesis – Local adaptation – Aphid performance Supervisors & advisors Martijn Bezemer Jeff Harvey Arjen Biere Tibor Bukovinszky Rieta Gols Thesis (major and minor) submitted in partial fulfillment of the requirements for the degree of Master of Sciences with the specialization "plant pathology and entomology". Research subject was originally proposed by Tibor Bukovinszky, Netherlands Institute for ecology (NIOO). Experimentations were done at the NIOO, Heteren, Netherlands, under the supervision of Arjen Biere, Jeff Harvey and Martijn Bezemer. Thesis process was supervised