Molecular Analysis of Sorbus Sp. from the Pieniny Mts. and Its Relation to Other Sorbus Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SZENT ISTVÁN EGYETEM Kertészettudományi Kar
SZENT ISTVÁN EGYETEM Kertészettudományi Kar SORBUS FAJKELETKEZÉS TRIPARENTÁLIS HIBRIDIZÁCIÓVAL A KELET- ÉS DÉLKELET- EURÓPAI TÉRSÉGBEN (Nothosubgenus Triparens) Doktori (PhD) értekezés Németh Csaba BUDAPEST 2019 A doktori iskola megnevezése: Kertészettudományi Doktori Iskola tudományága: Növénytermesztési és kertészeti tudományok vezetője: Zámboriné Dr. Németh Éva egyetemi tanár, DSc Szent István Egyetem, Kertészettudományi Kar, Gyógy- és Aromanövények Tanszék Témavezető: Dr. Höhn Mária egyetemi docens, CSc Szent István Egyetem, Kertészettudományi Kar, Növénytani Tanszék és Soroksári Botanikus Kert A jelölt a Szent István Egyetem Doktori Szabályzatában előírt valamennyi feltételnek eleget tett, az értekezés műhelyvitájában elhangzott észrevételeket és javaslatokat az értekezés átdolgozásakor figyelembe vette, azért az értekezés védési eljárásra bocsátható. .................................................. .................................................. Az iskolavezető jóváhagyása A témavezető jóváhagyása 2 Édesanyám emlékének. 3 4 TARTALOMJEGYZÉK RÖVIDÍTÉSEK JEGYZÉKE .......................................................................................................... 7 1. BEVEZETÉS ÉS CÉLKITŰZÉS .................................................................................................. 9 2. IRODALMI ÁTTEKINTÉS ..................................................................................................... 11 2.1. A Sorbus nemzetség taxonómiai vonatkozásai .................................................................... -

Biodiversität Von Schmetterlingen (Lepidoptera) Im Gebiet Des Naturparks Schlern
Gredleriana Vol. 7 / 2007 pp. 233 - 306 Biodiversität von Schmetterlingen (Lepidoptera) im Gebiet des Naturparks Schlern Peter Huemer Abstract Biodiversity of butterflies and moths (Lepidoptera) of the Schlern nature park (South Tyrol, Italy) The species diversity of Lepidoptera within the Schlern nature park has been explored during the vegetation periods 2006 and 2007. Altogether 1030 species have been observed in 16 sites, covering about one third of the entire fauna from the province of South Tyrol. Fifteen additional species have been collected during the diversity day 2006 whereas further 113 species date back to historical explorations. The species inventory includes 20 new records for South Tyrol which are briefly reviewed. Among these speciesMicropterix osthelderi, Rhigognostis incarnatella and Cydia cognatana are the first proved records from Italy. Further two species from the surroundings of the Park include the first published records ofPhyllonorycter issikii and Gelechia sestertiella for Italy. Site specific characteristics of Lepidoptera coenosis are discussed in some detail. Historical aspects of lepidopterological exploration of the Schlern are briefly introduced. Keywords: Lepidoptera, faunistics, biodiversity, Schlern, South Tyrol, Italy 1. Einleitung Der Schlern übt als einer der klassischen Dolomitengipfel schon weit über 100 Jahre eine geradezu magische Anziehungskraft auf Naturwissenschaftler unterschiedlichster Coleurs aus (Abb. 1). Seine Nähe zum Siedlungsraum und vor allem die damit verbundene relativ leichte Erreichbarkeit haben wohl zur Motivation mehrerer Forschergenerationen beigetragen, das Gebiet näher zu untersuchen. Zusätzlich war wohl die Arbeit von GREDLER (1863) über Bad Ratzes für manchen Entomologen ein Anlass die Fauna des Schlerns näher unter die Lupe zu nehmen. Dies gilt auch für Schmetterlinge, wo erste Aufsammlungen bereits in die Mitte des 19. -

Draft Carpathian Red List of Forest Habitats
CARPATHIAN RED LIST OF FOREST HABITATS AND SPECIES CARPATHIAN LIST OF INVASIVE ALIEN SPECIES (DRAFT) PUBLISHED BY THE STATE NATURE CONSERVANCY OF THE SLOVAK REPUBLIC 2014 zzbornik_cervenebornik_cervene zzoznamy.inddoznamy.indd 1 227.8.20147.8.2014 222:36:052:36:05 © Štátna ochrana prírody Slovenskej republiky, 2014 Editor: Ján Kadlečík Available from: Štátna ochrana prírody SR Tajovského 28B 974 01 Banská Bystrica Slovakia ISBN 978-80-89310-81-4 Program švajčiarsko-slovenskej spolupráce Swiss-Slovak Cooperation Programme Slovenská republika This publication was elaborated within BioREGIO Carpathians project supported by South East Europe Programme and was fi nanced by a Swiss-Slovak project supported by the Swiss Contribution to the enlarged European Union and Carpathian Wetlands Initiative. zzbornik_cervenebornik_cervene zzoznamy.inddoznamy.indd 2 115.9.20145.9.2014 223:10:123:10:12 Table of contents Draft Red Lists of Threatened Carpathian Habitats and Species and Carpathian List of Invasive Alien Species . 5 Draft Carpathian Red List of Forest Habitats . 20 Red List of Vascular Plants of the Carpathians . 44 Draft Carpathian Red List of Molluscs (Mollusca) . 106 Red List of Spiders (Araneae) of the Carpathian Mts. 118 Draft Red List of Dragonfl ies (Odonata) of the Carpathians . 172 Red List of Grasshoppers, Bush-crickets and Crickets (Orthoptera) of the Carpathian Mountains . 186 Draft Red List of Butterfl ies (Lepidoptera: Papilionoidea) of the Carpathian Mts. 200 Draft Carpathian Red List of Fish and Lamprey Species . 203 Draft Carpathian Red List of Threatened Amphibians (Lissamphibia) . 209 Draft Carpathian Red List of Threatened Reptiles (Reptilia) . 214 Draft Carpathian Red List of Birds (Aves). 217 Draft Carpathian Red List of Threatened Mammals (Mammalia) . -

Isoprenoid Emission in Hygrophyte and Xerophyte European Woody Flora: Ecological and Evolutionary Implications
Global Ecology and Biogeography, (Global Ecol. Biogeogr.) (2014) 23, 334–345 bs_bs_banner RESEARCH Isoprenoid emission in hygrophyte and PAPER xerophyte European woody flora: ecological and evolutionary implications Francesco Loreto1*, Francesca Bagnoli2, Carlo Calfapietra3,4, Donata Cafasso5, Manuela De Lillis1, Goffredo Filibeck6, Silvia Fineschi2, Gabriele Guidolotti7, Gábor Sramkó8, Jácint Tökölyi9 and Carlo Ricotta10 1Dipartimento di Scienze Bio-Agroalimentari, ABSTRACT Consiglio Nazionale delle Ricerche, Piazzale Aim The relationship between isoprenoid emission and hygrophily was investi- Aldo Moro 7, 00185 Roma, Italy, 2Istituto per la Protezione delle Piante, Consiglio Nazionale gated in woody plants of the Italian flora, which is representative of European delle Ricerche, Via Madonna del Piano 10, diversity. 50019 Sesto Fiorentino (Firenze), Italy, Methods Volatile isoprenoids (isoprene and monoterpenes) were measured, or 3 Istituto di Biologia Agroambientale e data collected from the literature, for 154 species native or endemic to the Medi- Forestale, Consiglio Nazionale delle Ricerche, terranean. The Ellenberg indicator value for moisture (EIVM) was used to describe Via Marconi 3, Porano (Terni), Italy, plant hygrophily. Phylogenetic analysis was carried out at a broader taxonomic scale 4Global Change Research Centre – CzechGlobe, on 128 species, and then refined on strong isoprene emitters (Salix and Populus Belidla 4a, 603 00 Brno, Czech Republic, species) based on isoprene synthase gene sequences (IspS). 5Dipartimento di Biologia, Università degli Studi di Napoli ‘Federico II, Complesso Results Isoprene emitters were significantly more common and isoprene emis- Universitario di Monte S. Angelo, Via Cinthia, sion was higher in hygrophilous EIVM classes, whereas monoterpene emitters were 80126 Napoli, Italy, 6Dipartimento di Scienze more widespread and monoterpene emission was higher in xeric classes. -

The Wild Food (Plants and Insects) in Western Friuli Local Knowledge (Friuli- Venezia Giulia, North Eastern Italy)
The wild food (plants and insects) in Western Friuli local knowledge (Friuli- Venezia Giulia, North Eastern Italy) a ngelo l. Dreon & Maurizio G. Paoletti ABSTRACt Contrib. Nat. Hist. 12: 461–488. Folk traditional diets in Western Friuli include wild plants and few invertebrates (i.e. insects) that are peculiar in the southern alps. 156 plants are listed here, ranging from 200 m to 2200 m a.s.l.. this selection is the result of the research of 64 inform- ants who contributed with their personal experiences. recovery of scattered know- ledge is valuable for maintaining and sustaining its use and enhancing a wild bio- diversity base for agroecology, conservation and ecotourism. Pistiç, frita and lidùm are some of the names used for about 50–60 herbs collected in field margins, hay meadows, woodlands and the wild, most commonly in spring. in order to be con- sumed, most herbs are boiled together and are later sautéed with butter or lard and garlic. the higher range pre-alpine Carnic slopes have supported seasonal sheep and cow grazing and summer pastoral communities have traditionally used plants as well for salads, soups and spices in addition to the boiled-sautéed dish. Furthermore, herbs have been used as medicinal elixirs and for milk processing. For instance, Lycopodium annotinum l. was employed to filter milk; Asplenium ruta-muraria l. was cooked with maize flour to prepare "fregole" a dish eaten with milk or coffee made from barley. specifically, collections of these useful alpine plants include: Cheno- podium bonus-henricus l., Aruncus dioicus (Walter) Fernald, Cicerbita alpina (l.) Wallr., Rumex alpinus l., Carlina acaulis l., Myrrhis odorata (l.) Scop., Ranunculus hybridus Biria (dry leaves used as pepper), and raw bulbils of Polygonum viviparum l. -

High Mountain Vegetation of the Apennines 179-194 10.Blasi Litt-Neu.Qxd 18.10.12 13:59 Seite 179
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Berichte der Reinhold-Tüxen-Gesellschaft Jahr/Year: 2012 Band/Volume: 24 Autor(en)/Author(s): Blasi Carlo, Vico Eva Del Artikel/Article: High mountain vegetation of the Apennines 179-194 10.Blasi_Litt-Neu.qxd 18.10.12 13:59 Seite 179 ©Reinhold-Tüxen-Gesellschaft (http://www.reinhold-tuexen-gesellschaft.de/) Ber. d. Reinh.-Tüxen-Ges. 24,179-194. Hannover 2012 High mountain vegetation of the Apennines – Carlo Blasi & Eva Del Vico, Rom – Abstract This paper outlines the more significant plant communities of the subalpine and alpine belts in the Apennines (shrub formations of the subalpine belt, alpine grasslands, snowbed vegetation, scree and rock fissure vegetation). This overview highlights the coenological autonomy of the Apennine vegetation from the communities of the Alps and central European mountains, as well as the similarities with the high-mountain vegetation of South-Eastern Europe. The paper emphasises the biogeographical value of the Apennines, with its unique biogeographic pattern especially in the central Apennines, the high conservation value in terms of flora, vegetation and habitats, and the key role for monitoring climate change. 1. Introduction The high altitude vegetation of the Apennines has been the subject of numerous studies (e.g. BIONDI et al. 1999, 2000, 2006; BLASI et al. 1989, 1990, 2003, 2005; DI PIETRO et al. 2008; DI PIETRO 2010; PETRAGLIA & TOMASELLI 2007; TOMASELLI 1994; TOMASELLI & ROSSI 1994; TOMASELLI et al. 2003, 2007), most of which have focused on the syntaxo- nomic features of these mountains. This paper does not focus on the syntaxonomy and tax- onomy of the Apennine chain, but refers to the plant sociological studies with the aim to high- light its great biogeographic and conservation importance (BLASI et al. -

Vocne Vrste U Pejzaznom Projektovanju.Pdf
ВОЋНЕ ВРСТЕ У ПЕЈЗАЖНОМ ПРОЈЕКТОВАЊУ доц. др Мирјана Љубојевић проф. др Владислав Огњанов дипл. инж. – мастер Ивана Сентић дипл. инж. – мастер Јована Дулић УНИВЕРЗИТЕТ У НОВОМ САДУ ПОЉОПРИВРЕДНИ ФАКУЛТЕТ Нови Сад, 2018. ЕДИЦИЈА „ОСНОВНИ УЏБЕНИК” Оснивач и издавач Едиције Универзитет у Новом Саду, Пољопривредни факултет Трг Доситеја Обрадовића 8, 21000 Нови Сад Година оснивања 1954. Главни и одговорни уредник едиције др Недељко Тица, редовни професор декан Пољопривредног факултета Чланови комисије за издавачку делатност др Љиљана Нешић, ванредни професор – председник др Бранислав Влаховић, редовни професор – члан др Милица Рајић, редовни професор – члан др Нада Плавша, ванредни професор – члан iii Аутори др Мирјана Љубојевић, доцент др Владислав Огњанов, редовни професор дипл. инж. – мастер Ивана Сентић, асистент дипл. инж. – мастер Јована Дулић, истраживач сарадник Главни и одговорни уредник др Недељко Тица, редовни професор декан Пољопривредног факултета у Новом Саду Уредник др Драгослав Иванишевић, ванредни професор, директор Департмана за воћарство, виноградарство, хортикултуру и пејзажну архитектуру Лектор Милица Митрушић професор српског језика и књижевности Рецензенти др Сандра Бијелић, ванредни професор др Милена Лакићевић, доцент Универзитет у Новом Саду, Пољопривредни факултет др Небојша Милошевић, научни сарадник Институт за воћарство, Чачак Издавач Универзитет у Новом Саду, Пољопривредни факултет, Нови Сад Забрањено прештампавање и фотокопирање. Сва права задржава издавач. Штампа: Штампање овог уџбеника одобрило је Наставно-научно веће Пољопривредног факултета у Новом Саду на седници од 18.12.2018. године. Број одлуке: 1000/0102 1436/1 Тираж: 20 Место и година штампања: 2018 iv Предговор Уџбеник „Воћне врсте у пејзажном пројектовању“ представља основно наставно средство за стицање знања и припрему испита из истоименог предмета који слушају студенти треће године студијских програма Хортикултура и пејзажна архитектура, док као помоћно средство може користити студентима студијског програма за воћарство и виноградарство. -
Spatial Structure of a Natural Mixed Topodeme of Subalpine Sorbus Taxa
Vol. 77, No. 4: 305-311, 2008 ACTA SOCIETATIS BOTANICORUM POLONIAE 305 SPATIAL STRUCTURE OF A NATURAL MIXED TOPODEME OF SUBALPINE SORBUS TAXA DUAN GÖMÖRY, DIANA KRAJMEROVÁ Technical University in Zvolen, Faculty of Forestry TG Masaryka 24, SK-960 53 Zvolen, Slovakia e-mail: [email protected] (Received: August 6, 2007. Accepted: March 20, 2008) ABSTRACT Spatial distribution and genetic variation of a population of Sorbus chamaemespilus (L.) Crantz and putative hybrids between S. chamaemespilus, S. aria and S. aucuparia growing in the nature reserve Skalná Alpa (central Slovakia) were studied. The analysis of spatial patterns using Ripleys K-function revealed a significant clustering of the adults of both S. chamaemespilus and hybrid taxa at distances up to ~15 m and a strong affinity between both taxonomical groups, indicating similar ecological requirements. Bivariate point-pattern analysis considering cardinal direction showed that juvenile individuals of S. chamaemespilus are clustered around the adults up to the distance of ~2 m, whereas in hybrid taxa with larger and more dense crowns, juveniles are clustered at distances more than ~3 m from the adults. The analysis of genetic variation in a subset of adult shrubs using 4 nuclear mi- crosatellite loci revealed that unlike expected, there was no variation in S. chamaemespilus but several genotypes were found in the group of hybrid taxa. Implications for the reproduction system and conservation of the investi- gated taxa are discussed. KEY WORDS: Sorbus L., spatial structure, apomixis, nuclear microsatellites, dispersal. INTRODUCTION tal taxa. Generally, they are limited to the subalpine zone of high mountain ranges in the central part of Western Car- The genus Sorbus L., just like the whole family Rosace- pathians. -

Sorbus (Rosaceae) in Herbarium of the Rechinger Family
ISSN 1211-8788 Acta Musei Moraviae, Scientiae biologicae (Brno) 89: 193–201, 2004 Sorbus (Rosaceae) in herbarium of the Rechinger family MILOSLAV KOVANDA Botanical Institute, Czech Academy of Sciences, 252 43 Prùhonice, Czech Republic Dedicated to the memory of Professor Karl Heinz Rechinger (1906–1998). KOVANDA M. 2004: Sorbus (Rosaceae) in herbarium of the Rechinger family. Acta Musei Moraviae, Scientiae biologicae (Brno) 89: 193–201. – A revision of Sorbus in the Rechinger family herbarium revealed the presence of S. aria (L.) Crantz, S. danubialis (Jáv.) Prodan, S. pannonica Kárpáti, S. chamaemespilus (L.) Crantz, S. torminalis (L.) Crantz, S. aucuparia L., S. domestica L. and a number of hybrids of S. aria × S. torminalis, S. aria × S. aucuparia and S. aria × S. chamaemespilus parentage. The results are briefly discussed. Key words. Rosaceae, Sorbus, Rechinger K. H., herbarium Introduction Shortly before his death in December 1998, Professor Karl Heinz Rechinger, a renowned authority on the flora of the Balkan Peninsula and S.W. Asia, suggested in a friendly letter that I should inspect the Sorbus material in his family herbarium. He arranged for the material, which is kept at the Conservatoire et Jardin Botaniques in Geneva, to be sent to me on loan. The consignment contained 561 sheets, excellently prepared and labelled, originating mostly from central European countries: Austria (all federal states – “Bundesländer” – except Vorarlberg), Hungary, the Czech Republic, Slovakia, Germany, northern Italy, and the Balkan Peninsula – Croatia, Serbia, Bosnia and Herzegovina, Yugoslavia, Macedonia, Rumania, Bulgaria and Greece. A few collections originated from botanical gardens in Sweden. The vast majority of the specimens were collected by members of the Rechinger family (Karl Rechinger, Lily Rechinger and Karl Heinz Rechinger) but important additions came from J. -
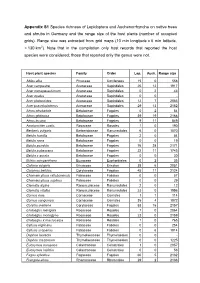
Appendix S1 Species Richness of Lepidoptera and Auchenorrhyncha on Native Trees and Shrubs in Germany and the Range Size of the Host Plants (Number of Occupied Grids)
Appendix S1 Species richness of Lepidoptera and Auchenorrhyncha on native trees and shrubs in Germany and the range size of the host plants (number of occupied grids). Range size was extracted from grid maps (10 min longitude x 6 min latitude, ≈ 130 km 2). Note that in the compilation only host records that reported the host species were considered; those that reported only the genus were not. Host plant species Family Order Lep. Auch. Range size Abies alba Pinaceae Coniferales 15 0 558 Acer campestre Aceraceae Sapindales 25 12 1917 Acer monspessulanum Aceraceae Sapindales 0 2 43 Acer opalus Aceraceae Sapindales 0 0 - Acer platanoides Aceraceae Sapindales 12 7 2083 Acer pseudoplatanus Aceraceae Sapindales 29 13 2152 Alnus alnobetula Betulaceae Fagales 0 2 54 Alnus glutinosa Betulaceae Fagales 39 19 2168 Alnus incana Betulaceae Fagales 9 11 849 Amelanchier ovalis Rosaceae Rosales 1 0 190 Berberis vulgaris Berberidaceae Ranunculales 6 0 1070 Betula humilis Betulaceae Fagales 2 0 54 Betula nana Betulaceae Fagales 0 0 19 Betula pendula Betulaceae Fagales 76 28 2171 Betula pubescens Betulaceae Fagales 22 11 1745 Betula x aurata Betulaceae Fagales 0 0 30 Buxus sempervirens Buxaceae Euphorbiales 0 2 35 Calluna vulgaris Ericaceae Ericales 38 6 2051 Carpinus betulus Corylaceae Fagales 45 11 2124 Chamaecytisus ratisbonensis Fabaceae Fabales 0 0 57 Chamaecytisus supinus Fabaceae Fabales 0 0 29 Clematis alpina Ranunculaceae Ranunculales 2 0 12 Clematis vitalba Ranunculaceae Ranunculales 23 0 1556 Cornus mas Cornaceae Cornales 1 1 114 Cornus sanguinea -

The State of the World's Forest Genetic Resources: Country Report
SWITZERLAND This country report is prepared as a contribution to the FAO publication, The Report on the State of the World’s Forest Genetic Resources. The content and the structure are in accordance with the recommendations and guidelines given by FAO in the document Guidelines for Preparation of Country Reports for the State of the World’s Forest Genetic Resources (2010). These guidelines set out recommendations for the objective, scope and structure of the country reports. Countries were requested to consider the current state of knowledge of forest genetic diversity, including: Between and within species diversity List of priority species; their roles and values and importance List of threatened/endangered species Threats, opportunities and challenges for the conservation, use and development of forest genetic resources These reports were submitted to FAO as official government documents. The report is presented on www. fao.org/documents as supportive and contextual information to be used in conjunction with other documentation on world forest genetic resources. The content and the views expressed in this report are the responsibility of the entity submitting the report to FAO. FAO may not be held responsible for the use which may be made of the information contained in this report. Final FGR2012_CountryReport_Switzerland_3 5 docx 1 / 52 THE STATE OF THE WORLD’S FOREST GENETIC RESSOURCES COUNTRY REPORT SWITZERLAND 2012 ETHZ/BAFU 10.03.2013 Final FGR2012_CountryReport_Switzerland_3 5 docx 2 / 52 IMPRESSUM The present report was prepared -

F2.4 Subalpine Pinus Mugo Scrub
European Red List of Habitats - Heathland Habitat Group F2.4 Subalpine Pinus mugo scrub Summary Conifer krummholz scrub dominated by Pinus mugo occurs above the timberline and on subalpine screes in the mountains of central and southeastern Europe, on both calcareous and siliceous bedrock. It can be short or tall, closed or open and other shrubs, sub-shrubs and herbaceous associates vary according to soil acidity and wetness. Recovery after periodic burning can be rapid and the scrub can spread into abandoned pastures but a lasting threat is clearance for tourist developments. Careful planning and avoiding burning are the best conservation measures. Synthesis The Red List criteria qualify this habitat for a Least Concern (LC) status as there is only a small negative trend in quantity and in quality over the last 50 years, and the habitat is relative widely distributed. Overall Category & Criteria EU 28 EU 28+ Red List Category Red List Criteria Red List Category Red List Criteria Least Concern - Least Concern - Sub-habitat types that may require further examination No sub-habitats need to be distinguished for further analysis. Habitat Type Code and name F2.4 Subalpine Pinus mugo scrub Extensive stands of the Pinus mugo scrub in a glacial cirque in the Retezat Mts, Pinus mugo scrub on the summit of Mt Grosser Arber, Bavarian Forest, Germany Romania (Photo: Milan Chytrý) (Photo: Milan Chytrý). Habitat description Conifer scrub dominated by Pinus mugo (krummholz) occurring in the mountains of central and southeastern Europe above the timberline. This scrub is usually 0.5-3 m tall, depending on the wind exposure of the site and the height of winter snow cover.