Unveiling New Disease, Pathway, and Gene Associations Via Multi-Scale Neural Networks
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NCI RAS Initiative Update
Ras Initiative Update Frank McCormick and Levi Garraway DEPARTMENT OF HEALTH AND HUMAN SERVICES • National Institutes of Health • National Cancer Institute The Frederick National Laboratory is a Federally Funded Research and Development Center operated by Leidos Biomedical Research, Inc., for the National Cancer Institute RAS Initiative Accomplishments: Evaluating Ras dependency 2 SiREN assay for Ras dependency SiREN assay for Ras dependency 70% KD siEGFP 0-10% KD 50% KD 100% KD SiREN assay for Ras dependency RAS Initiative Accomplishments: Biophysical and structural analysis 7 Ras proteins 1 -166 167-185,6 Raf, PI 3’ kinase RalGDS, GAPs Raf, PI 3’ kinase RalGDS, GAPs Raf, PI 3’ kinase RalGDS, GAPs Raf, PI 3’ kinase RalGDS, GAPs Palmitoyl Farnesyl Fully processed KRAS4b A. Gorfe, U-Texas Houston Engineering baculovirus for improved production of processed KRAS • recombineering used to insert FNTA/FNTB genes into the baculovirus genome • eliminated issues with coinfection of multiple viruses • maltose-binding protein (MBP) fusion for greater yield and solubility • Trichoplusia ni (Hi5) insect cells for increased yield Carissa Grose, Dom Esposito, Bill Gillette Processed KRAS4b characterization • Extensive protein characterization – Purified to homogeneity; yield >7mg/L – Intact mass – Predominantly monomeric – Secondary structure equivalent to non-processed KRAS4b KRAS4b- FME – Lower thermal stability Intact mass analysis Analytical ultracentrifugation Secondary structure by CD 2.14 S 25.7 kDa 10 Bill Gillette, Zhaojing Meng, Shelley Perkins, -

Miriam Molina Arcas
CV Miriam Molina Arcas Miriam Molina Arcas Principal Laboratory Research Scientist The Francis Crick Institute 1 Midland Road, NW1 1AT London +442037963313 [email protected] EDUCATION 1999-2005 PhD in Biomedicine, University of Barcelona, Spain Role of equilibrative nucleoside transporters in the sensitivity to antineoplasic drugs. Supervisors: Prof. Marçal Pastor-Anglada and Dr. F. Javier Casado 1994-1999 BSc Biochemistry, University of Barcelona, Spain RESEARCH AND PROFESSIONAL EXPERIENCE June 2016- present Senior Laboratory Research Scientist, Oncogene Biology Laboratory, The Francis Crick Institute. Prof. Julian Downward. Studied combination approaches that improve the efficacy of KRAS inhibitors and reduce resistance mechanisms. March 2015- May 2016 Postdoctoral Fellow, Lung Cancer Team, Institute of Cancer Research. Prof. Julian Downward. Designed and optimized a whole-genome shRNA screen to identify synthetic lethal interactions with MEK and IGF1R inhibitors. Sep 2008- Feb 2015 Postdoctoral Fellow, Signal Transduction Laboratory, Cancer Research UK- London Research Institute. Prof. Julian Downward Using RNA interfering and drug screens, identified new therapeutic strategies to treat tumours harbouring KRAS mutations 2006- Aug 2008 Postdoctoral Fellow, University of Barcelona, Spain Prof. Marçal Pastor-Anglada Studied the role of membrane transporters in nucleoside-derived drug response in cancer and HIV. TEACHING AND MENTORING EXPERIENCE 2018-2021 Honorary Research Associate in the School of Life & Medical Sciences, University College London, part of the Certificate in Core Teaching Practices. 2007-2008 Part-time lecturer of Biochemistry and Molecular Biology (University of Barcelona). 180h of theory and practical lessons. 2003-2005 Teaching assistant of Biochemistry and Molecular Biology (University of Barcelona). 60h of practical lessons. September 2019 Co-supervisor of MSc Experimental Pharmacology and Therapeutics Research of Pablo Romero Clavijo at the University College London. -

Fellows Opted out of the Mentoring Programme As of 21 July 2020 1 of 3 Professor Chris Abell FRS Fmedsci Professor Philip Jones
Fellows opted out of the Mentoring programme as of 21 July 2020 Professor Chris Abell FRS FMedSci Professor Philip Jones FMedSci Professor David Adams FMedSci Professor Roger Jones FMedSci Professor Judith Allen FRSE FMedSci Professor Kamlesh Khunti FMedSci Professor Thomas Kirkwood CBE Professor Desmond Archer OBE FMedSci FMedSci Professor Louise Arseneault FMedSci Professor Robb Krumlauf FMedSci Professor Charalambos Kyriacou Professor Michael Arthur FMedSci FMedSci Professor Deborah Ashby OBE FMedSci Sir Peter Lachmann FRS FMedSci Professor Dame Frances Ashcroft DBE Professor Leon Lagnado FMedSci FRS FMedSci Professor Anthony Barrett FRS FMedSci Professor Ajit Lalvani FMedSci Professor Gillian Bates FRS FMedSci Professor Deborah Lawlor CBE FMedSci Professor Facundo Batista FMedSci Professor Joy Lawn FMedSci Professor Stephan Beck FMedSci Professor Susan Lea FMedSci Professor Jill Belch OBE FMedSci Professor Sir Robert Lechler PMedSci Professor Sir John Bell GBE FRS Dr Melanie Lee CBE FMedSci HonFREng FMedSci Professor Wendy Bickmore FRS FRSE Professor Andrew Lees FMedSci FMedSci Professor Sir Adrian Bird CBE FRS FRSE Professor Stafford Lightman FRS FMedSci FMedSci Professor Ewan Birney CBE FRS Professor Janet Lord FMedSci FMedSci Professor Dorothy Bishop FRS FBA Professor Thomas MacDonald FMedSci FMedSci Professor Rona MacKie CBE FRSE Professor Jane Blazeby FMedSci Sir Stephen Bloom FRS FMedSci Professor Giovanna Mallucci FMedSci Sir Tom Blundell FRS FMedSci Dr Fiona Marshall FMedSci Professor Dame Theresa Marteau DBE Professor Peter -

Drugging the Undruggable: Advances on RAS Targeting in Cancer
G C A T T A C G G C A T genes Review Drugging the Undruggable: Advances on RAS Targeting in Cancer Miriam Molina-Arcas 1,*, Amit Samani 1,2,* and Julian Downward 1,3 1 Oncogene Biology Laboratory, Francis Crick Institute, London NW1 1AT, UK; [email protected] 2 Department of Medical Oncology, Imperial College Healthcare NHS Trust, London W2 1NY, UK 3 Lung Cancer Group, Institute of Cancer Research, London SW3 6JB, UK * Correspondence: [email protected] (M.M.-A.); [email protected] (A.S.) Abstract: Around 20% of all malignancies harbour activating mutations in RAS isoforms. Despite this, there is a deficiency of RAS-targeting agents licensed for therapeutic use. The picomolar affinity of RAS for GTP, and the lack of suitable pockets for high-affinity small-molecule binding, precluded effective therapies despite decades of research. Recently, characterisation of the biochemical properties of KRAS-G12C along with discovery of its ‘switch-II pocket’ have allowed development of effective mutant-specific inhibitors. Currently seven KRAS-G12C inhibitors are in clinical trials and sotorasib has become the first one to be granted FDA approval. Here, we discuss historical efforts to target RAS directly and approaches to target RAS effector signalling, including combinations that overcome limitations of single-agent targeting. We also review pre-clinical and clinical evidence for the efficacy of KRAS-G12C inhibitor monotherapy followed by an illustration of combination therapies designed to overcome primary resistance and extend durability of response. Finally, we briefly discuss novel approaches to targeting non-G12C mutant isoforms. -

Second RAS Initiative Symposium Sponsored by the National Cancer Institute Advanced Technology Research Facility, Frederick, MD December 6-8, 2017
The Second RAS Initiative Symposium Sponsored by the National Cancer Institute Advanced Technology Research Facility, Frederick, MD December 6-8, 2017 December 6 8:00 am – 8:15 am Welcome from the NCI Director Norman Sharpless 8:15 am – 9:00 am Keynote Frank McCormick (RAS Initiative, University of California, San Francisco) 9:00 am – 9:30 am Organoids as Model Systems for Tumorigenesis Calvin Kuo (Stanford University) 9:30 am – 9:50 am Break 9:50 am – 12:10 pm Activated RAS and Oncogenesis - Chair: Jeff Settleman Douglas Lowy (National Cancer Institute) Christopher Counter (Duke University) Matthew Vander Heiden (Massachusetts Institute of Technology) Marie Evangelista (Genentech) Aaron Hobbs (Der Lab, University of North Carolina, Chapel Hill) 12:10 pm – 1:10 pm Lunch on your own 1:10 pm – 3:15 pm Poster Session #1 3:15 pm – 4:15 pm RAS Multimers at the Membrane - Chair: John Hancock Yao-Cheng Li (Wahl Lab, Salk Institute) Margie Sutton (Bast Lab, MD Anderson) Chiara Ambrogio (Jänne Lab, Dana Farber Cancer Institute) 4:15 pm – 5:30 pm RAS Membrane Modeling and Dynamics - Chair: Dwight Nissley Roland Winter (Technische Universität Dortmund) Fred Streitz (Lawrence Livermore National Laboratory) Chris Neale (Garcia Lab, Los Alamos National Laboratory) Walkersville Social Hall 79 West Frederick Street, Walkersville 6:15 – 7:00 pm Hors d’oeuvres, cash bar 7:00 pm Dinner December 7 8:00 am – 10:00 am Tumor Dynamics - Chair: Rosalie Sears Dafna Bar-Sagi (New York University) David Tuveson (Cold Spring Harbor Laboratory) Channing Der (University -

Combining Three Antibodies Nullifies Feedback-Mediated Resistance to Erlotinib in Lung Cancer
RESEARCH ARTICLE CANCER Combining three antibodies nullifies feedback-mediated resistance to erlotinib in lung cancer Maicol Mancini,1 Nadège Gaborit,1 Moshit Lindzen,1 Tomer Meir Salame,2 Massimiliano Dall’Ora,1 Michal Sevilla-Sharon,1 Ali Abdul-Hai,1,3 Julian Downward,4,5 Yosef Yarden1* Despite initial responses to targeted kinase inhibitors, lung cancer patients presenting with primary epidermal growth factor receptor (EGFR) mutations acquire resistance, often due to a second-site mutation (T790M). However, clinical trials found no survival benefits in patients treated with a monoclonal antibody (mAb) to EGFR that should block activation of the mutated receptor and thus bypass resistance to molecules that target the catalytic or ATP-binding site. Using cell lines with the T790M mutation, we discovered that pro- longed exposure to mAbs against only the EGFR triggered network rewiring by (i) stimulating the extra- Downloaded from cellular signal–regulated kinase (ERK) pathway; (ii) inducing the transcription of HER2 (human epidermal growth factor receptor 2) and HER3, which encode other members of the EGFR family, and the gene encoding HGF, which is the ligand for the receptor tyrosine kinase MET; and (iii) stimulating the interaction between MET and HER3, which promoted MET activity. Supplementing the EGFR-specific mAb with those targeting HER2 and HER3 suppressed these compensatory feedback loops in cultured lung cancer cells. The triple mAb com- bination targeting all three receptors prevented the activation of ERK, accelerated the degradation of the re- ceptors, inhibited the proliferation of tumor cells but not of normal cells, and markedly reduced the growth of http://stke.sciencemag.org/ tumors in mice xenografted with cells that were resistant to combined treatment with erlotinib and the single function-blocking EGFR mAb. -
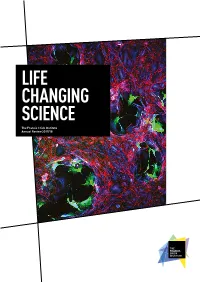
Life Changing Science
LIFE CHANGING SCIENCE The Francis Crick Institute Annual Review 2017/18 AN INSTITUTE FOR DISCOVERY Our commitment to excellence, our emphasis on multidisciplinary research, our focus on young and emerging talent and our novel ways of partnership working are some of the factors that set the Crick apart. Front cover Vaccinia virus infection (green) disrupts a layer of epithelial cells (red/blue). Courtesy of Michael Way, Group Leader at the Crick. INTRODUCTION 2 Who we are Our year at a glance 2 Introduction by Paul Nurse 4 The Francis Crick Institute is a biomedical Progress against our strategy 6 discovery institute dedicated to understanding the RESEARCH HIGHLIGHTS 10 Cancer-causing mutation fundamental biology underlying health and disease. suppresses immune system 11 Our work is helping to build an understanding of Predicting lung cancer’s return 12 New understanding of human why disease develops and to translate discoveries embryo development 14 Chemical attraction could improve into new ways to prevent, diagnose and treat cancer immunotherapy 16 illnesses such as cancer, heart disease, stroke, Genes linked to malaria parasites’ persistence 17 infections and neurodegenerative diseases. Architecture of our ‘second brain’ 18 Cause of infertility side-stepped in mice 19 Mechanism for spinal cord development discovered 20 A new layer of complexity in embryo development 21 Two DNAs wedded with this ring 22 Unravelling how DNA gets copied 23 Telomerase’s dark side discovered 24 REVIEW OF THE YEAR 26 New group leaders arrive 27 Joined-up thinking 30 Focusing on the molecules of life 32 CryoEM at the Crick 34 Bringing academia and industry closer together 36 The people making research happen 38 Patterns in art and science 40 Rewarding research 42 Appointments 43 Supporting new discoveries 44 Our vision What’s inside Our vision is to be a world- We bring together outstanding scientists Science feature 32 leading multidisciplinary from all disciplines and carry out research Sophisticated microscopy is being biomedical research institute. -

SCHEDULE of EVENTS Saturday, April 5 ALL SESSIONS ELIGIBLE for CME CREDIT UNLESS OTHERWISE NOTED.*
09_14AM_SchedEvents_Layout 1 3/11/14 12:29 PM Page 101 SCHEDULE OF EVENTS Saturday, April 5 ALL SESSIONS ELIGIBLE FOR CME CREDIT UNLESS OTHERWISE NOTED.* Saturday, April 5, 2014 Integrative Molecular Epidemiology , Thomas A. Sellers, Chairperson, Room 25, San Diego Convention Center, p. 146 8:00 a.m.-10:00 a.m. Educational Sessions Speakers: Thomas A. Sellers, Lorelei A. Mucci, Simon A. Gayther, Peter Kraft Beyond Ipilimumab and PD1/PD-L1 Pathway Blockade: Next Generation of Immunomodulatory PI3K-mTOR/PTEN , Ramon E. Parsons, Chairperson, Antibodies , Mario Sznol, Chairperson, Room 33, Room 31, San Diego Convention Center, p. 146 San Diego Convention Center, p. 143 Speakers: Lewis C. Cantley, Bart Vanhaesebroeck, John Speakers: Mario Sznol, Ana C. Anderson, Drew M. Blenis, Ramon E. Parsons Pardoll, Andrew D. Weinberg Targeting RhoGTPases and Their Kinase Effectors , Cancer Metabolism , Chi V. Dang, Chairperson, Jonathan Chernoff, Chairperson, Room 1, San Diego Room 6CF, San Diego Convention Center, p. 143 Convention Center, p. 146 Speakers: Chi V. Dang, Joshua D. Rabinowitz, Matthew Speakers: Jonathan Chernoff, Nupam P. Mahajan, G. Vander Heiden, Teresa W. M. Fan Cristina Fernandez-Valle, Michael F. Olson Drugging the Cell Cycle in Cancer , Geoffrey I. Shapiro, Tumor Immunology and Immunotherapy for Non- Chairperson, Room 7, San Diego Convention Center, Immunologists , Michael A. Caligiuri, Chairperson, p. 144 Ballroom 20D, San Diego Convention Center, p. 147 Speakers: Geoffrey I. Shapiro, David D. L. Bowtell, Alan Speakers: Robert A. Baiocchi, Lisa M. Coussens, Jeffrey Eastman, Wenyi Wei A. Sosman, Crystal L. Mackall Evolving Tumor Cell Populations: Dealing with Diversity , Nicholas E. Navin, Chairperson, Room 11, San Diego Convention Center, p. -

Kras and Tumor Immunity: Friend Or Foe?
Downloaded from http://perspectivesinmedicine.cshlp.org/ on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press Kras and Tumor Immunity: Friend or Foe? Jane Cullis,1 Shipra Das,1 and Dafna Bar-Sagi Department of Biochemistry and Molecular Pharmacology, New York University School of Medicine, New York, New York 10016 Correspondence: [email protected] With the recent breakthroughs in immunotherapy as curative treatments in certain tumor types, there has been renewed interest in the relationship between immunity and tumor growth. Although we are gaining a greater understanding of the complex interplay of immune modulating components in the tumor microenvironment, the specific role that tumor cells play in shaping the immune milieu is still not well characterized. In this review, we focus on how mutant Kras tumor cells contribute to tumor immunity, with a specific focus on processes induced directly or indirectly by the oncogene. t is well recognized that the growth of mutant ment and progression but also sensitivity to im- IKras tumors is associated with an immuno- munotherapy (Topalian et al. 2012; Pauken et al. suppressed state that is established via the 2015). In the sections that follow, we will elabo- dynamic interplay between components of rate on how mutant Kras-regulated signaling both the innate and adaptive immune response pathways affect the presence and function of (Fig. 1). For example, immunosuppressive cells these immune cell types. Moreover, we will commonly associated with mutant Kras tumors describe -

Michel Foucault Ronald C Kessler Graham Colditz Sigmund Freud
ANK RESEARCHER ORGANIZATION H INDEX CITATIONS 1 Michel Foucault Collège de France 296 1026230 2 Ronald C Kessler Harvard University 289 392494 3 Graham Colditz Washington University in St Louis 288 316548 4 Sigmund Freud University of Vienna 284 552109 Brigham and Women's Hospital 5 284 332728 JoAnn E Manson Harvard Medical School 6 Shizuo Akira Osaka University 276 362588 Centre de Sociologie Européenne; 7 274 771039 Pierre Bourdieu Collège de France Massachusetts Institute of Technology 8 273 308874 Robert Langer MIT 9 Eric Lander Broad Institute Harvard MIT 272 454569 10 Bert Vogelstein Johns Hopkins University 270 410260 Brigham and Women's Hospital 11 267 363862 Eugene Braunwald Harvard Medical School Ecole Polytechnique Fédérale de 12 264 364838 Michael Graetzel Lausanne 13 Frank B Hu Harvard University 256 307111 14 Yi Hwa Liu Yale University 255 332019 15 M A Caligiuri City of Hope National Medical Center 253 345173 16 Gordon Guyatt McMaster University 252 284725 17 Salim Yusuf McMaster University 250 357419 18 Michael Karin University of California San Diego 250 273000 Yale University; Howard Hughes 19 244 221895 Richard A Flavell Medical Institute 20 T W Robbins University of Cambridge 239 180615 21 Zhong Lin Wang Georgia Institute of Technology 238 234085 22 Martín Heidegger Universität Freiburg 234 335652 23 Paul M Ridker Harvard Medical School 234 318801 24 Daniel Levy National Institutes of Health NIH 232 286694 25 Guido Kroemer INSERM 231 240372 26 Steven A Rosenberg National Institutes of Health NIH 231 224154 Max Planck -

SCIENTIFIC REPORT 2017 Cruk.Org SCIENTIFIC COVER IMAGE Mitochondria, the Powerhouses of the Cell, Are Required for Cancer Cell Death
SCIENTIFIC REPORT 2017 cruk.org SCIENTIFIC COVER IMAGE Mitochondria, the powerhouses of the cell, are required for cancer cell death. This image shows mitochondrial shape REPORT changes during cancer cell death, where each colour represents mitochondrial shape at a specific time point during cell death. Image supplied by Joel Riley, a post-doc in Stephen Tait’s group. 2017 BEATSON INSTITUTE CONTENTS SECTION 1 Shehab Ismail 52 DIRECTOR'S INTRODUCTION 04 Structural Biology of Cilia INSTITUTE OF CANCER SCIENCES 06 Laura Machesky 54 INTRODUCTION Migration, Invasion and Metastasis RESEARCH HIGHLIGHTS 08 Jim Norman 56 GABRIELA KALNA OBITUARY 11 Integrin Cell Biology TESSA HOLYOAKE OBITUARY 12 Michael Olson 58 BACKGROUND 13 Molecular Cell Biology Owen Sansom 60 CANCER RESEARCH UK BEATSON INSTITUTE Colorectal Cancer and Wnt Signalling Sara Zanivan 62 CANCER GROWTH AND METABOLISM Tumour Microenvironment and Proteomics Imran Ahmad 16 Models of Advanced Prostate Cancer DRUG DISCOVERY Peter D. Adams 18 Justin Bower & Heather McKinnon 66 Epigenetics of Cancer and Ageing Drug Discovery Unit Eyal Gottlieb 20 Tumour Metabolism ADVANCED TECHNOLOGIES Danny Huang 22 Leo Carlin 70 Ubiquitin Signalling Beatson Advanced Imaging Resource (BAIR) Jurre Kamphorst 24 Bioinformatics and Computational Biology 72 Cancer Metabolomics Sara Zanivan 73 Hing Leung 26 Proteomics Prostate Cancer Biology Emma Shanks 74 David Lewis 28 Functional Screening Molecular Imaging Gillian Mackay 76 Daniel J. Murphy 30 Metabolomics Oncogene-Induced Vulnerabilities Jennifer Morton 78 -

Editor European Editor Senior Editors Consulting Editor Managing
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Cell Editor Associate Editors Benjamin Lewin Richard Axel Philippa Marrack Rosa Beddington Chris Marshall European Editor Pierre Chambon Diane Mathis Peter W. J. Rigby Joanne Chory Ira Mellman Julian Downward Don Metcalf Senior Editors David Friedman Elliot Meyerowitz Amy Axelrod Phillip Gallimore Tim Mitchison Vivian Siegel John Gerhart Paul Nurse Stephen Goff William Paul Joe Goldstein Hugh Pelham Consulting Editor Peter Goodfellow Roger Perlmutter Gregory Gasic Peter Gruss Klaus Rajewsky Martin Hemier Danny Reinberg Managing Editor Ira Herskowitz James Roberts Rebecca Kennison Chris Higgins Elliott Ross Robert Horvitz Gerald Rubin Editorial Staff Tim Hunt Joshua Sanes Alexander Barnett Tony Hunter Jeff Schell Edward J. Dionne Richard Hynes Matthew Scott Jennifer MacMillan David Ish-Horowitz David Sherratt Martha Sullivan Thomas Jesse11 Jim Smith Peter Kim Frank Solomon Judith Kimble Davor Solter Advertising Roger Kornberg George Sprague Lynn Reznick Parisi - Director John Kuriyan Charles Stevens Jennifer Oneglia - Classifieds Ron Laskey Robert Tjian Christa A. Matukaitis Michael Levine Richard Treisman Richard Losick Mike Waterfield Circulation Vivek Malhotra Alan Weiner Marie Arsenault - Manager Tom Maniatis Owen Witte Heather L. McCormick Telephone Numbers Principal Office European Office Cell Division of Eukaryotic Editorial: 617-661-7057 1050 Massachusetts Avenue Molecular Genetics Advertising: 617-661-7059 Cambridge, Massachusetts 02138 NIMR USA The Ridgeway, Mill Hill Subscriptions: 617-661-7060 Fax: 617-661-7061 London NW7 1 AA, England E-mail: [email protected] Telephone: (0)181-906-3897 Fax: (0)181-913-8527 E-mail: [email protected] Cell (ISSN 0092-8674) is published biweekly by Cell Press, 1050 Massachusetts Avenue, Cambridge, Massachusetts 02138.