Book Halim.Indb
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Exome Sequencing Identifies Mutations in the Gene TTC7A In
Developmental defects J Med Genet: first published as 10.1136/jmedgenet-2012-101483 on 19 February 2013. Downloaded from ORIGINAL ARTICLE Exome sequencing identifies mutations in the gene TTC7A in French-Canadian cases with hereditary multiple intestinal atresia Mark E Samuels,1 Jacek Majewski,2 Najmeh Alirezaie,2 Isabel Fernandez,1,3 Ferran Casals,1 Natalie Patey,1,4 Hélène Decaluwe,1,5 Isabelle Gosselin,6 Elie Haddad,1,3,5 Alan Hodgkinson,1 Youssef Idaghdour,1 Valerie Marchand,1,5 1,5 6,7 6 6 Open Access Jacques L Michaud, Marc-André Rodrigue, Sylvie Desjardins, Stéphane Dubois, Scan to access more 1,3 1,5 6,7 8 free content Francoise Le Deist, Philip Awadalla, Vincent Raymond, Bruno Maranda ▸ Additional material is ABSTRACT attempted, outcomes are poor and the condition is published online only. To view Background Congenital multiple intestinal atresia usual fatal within the first month of life. To date, please visit the journal online (http://dx.doi.org/10.1136/ (MIA) is a severe, fatal neonatal disorder, involving the no primary aetiology has been proved for the con- jmedgenet-2012-101483). occurrence of obstructions in the small and large dition. Importantly, in some cases MIA is also asso- 1 intestines ultimately leading to organ failure. Surgical ciated with either mild or severe combined Centre de Recherche du CHU fi 2–5 Ste-Justine, University of interventions are palliative but do not provide long-term immunode ciency (SCID), raising the possibility Montreal, Montreal, Quebec, survival. Severe immunodeficiency may be associated that an abnormal immune response might be the Canada with the phenotype. -
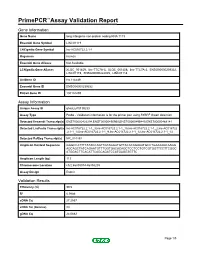
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name long intergenic non-protein coding RNA 1119 Ensembl Gene Symbol LINC01119 LNCipedia Gene Symbol lnc-AC016722.2.1-1 Organism Human Ensembl Gene Aliases Not Available LCNipedia Gene Aliases XLOC_001459, linc-TTC7A-3, XLOC_001458, linc-TTC7A-2, ENSG00000239332, LINC01119, ENSG00000222005, LINC01118 UniGene ID Hs.114449 Ensembl Gene ID ENSG00000239332 Entrez Gene ID 100134259 Assay Information Unique Assay ID qhsaLEP0139233 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Ensembl Transcript(s) ENST00000422294,ENST00000490950,ENST00000495449,ENST00000468141 Detected LncPedia Transcript(s) lnc-AC016722.2.1-1_3,lnc-AC016722.2.1-1_15,lnc-AC016722.2.1-1_2,lnc-AC016722 .2.1-1_14,lnc-AC016722.2.1-1_9,lnc-AC016722.2.1-1_12,lnc-AC016722.2.1-1_13 Detected RefSeq Transcript(s) NR_024452 Amplicon Context Sequence GGGCCATTTTATACCAGTTGTAGGATGTTACACAGAAATGCCTGAAAAGCAAGG AGCAGCTATCAGAATGTTTGGTGACACAGCTCCTCCTGTCGTGGTTCCTTCGGC ATGGACTTCACATTCAGCAGATCCATGAGGTGTTC Amplicon Length (bp) 113 Chromosome Location chr2:46855974-46858205 Assay Design Exonic Validation Results Efficiency (%) 99.5 R2 0.9986 cDNA Cq 27.2537 cDNA Tm (Celsius) 83 gDNA Cq 24.0682 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report LINC01119, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

TTC7A Mutations Disrupt Intestinal Epithelial Apicobasal Polarity
TTC7A mutations disrupt intestinal epithelial apicobasal polarity Amélie E. Bigorgne, … , Hans Clevers, Geneviève de Saint Basile J Clin Invest. 2014;124(1):328-337. https://doi.org/10.1172/JCI71471. Research Article Gastroenterology Multiple intestinal atresia (MIA) is a rare cause of bowel obstruction that is sometimes associated with a combined immunodeficiency (CID), leading to increased susceptibility to infections. The factors underlying this rare disease are poorly understood. We characterized the immunological and intestinal features of 6 unrelated MIA-CID patients. All patients displayed a profound, generalized lymphocytopenia, with few lymphocytes present in the lymph nodes. The thymus was hypoplastic and exhibited an abnormal distribution of epithelial cells. Patients also had profound disruption of the epithelial barrier along the entire gastrointestinal tract. Using linkage analysis and whole-exome sequencing, we identified 10 mutations in tetratricopeptide repeat domain–7A (TTC7A), all of which potentially abrogate TTC7A expression. Intestinal organoid cultures from patient biopsies displayed an inversion of apicobasal polarity of the epithelial cells that was normalized by pharmacological inhibition of Rho kinase. Our data indicate that TTC7A deficiency results in increased Rho kinase activity, which disrupts polarity, growth, and differentiation of intestinal epithelial cells, and which impairs immune cell homeostasis, thereby promoting MIA-CID development. Find the latest version: https://jci.me/71471/pdf Research article TTC7A mutations disrupt intestinal epithelial apicobasal polarity Amélie E. Bigorgne,1,2,3 Henner F. Farin,4 Roxane Lemoine,1,2,3 Nizar Mahlaoui,2,3 Nathalie Lambert,5 Marine Gil,5 Ansgar Schulz,6 Pierre Philippet,7 Patrick Schlesser,7 Tore G. Abrahamsen,8 Knut Oymar,9 E. -
![CD98 [19] Among Others [5][23]](https://docslib.b-cdn.net/cover/4111/cd98-19-among-others-5-23-1304111.webp)
CD98 [19] Among Others [5][23]
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.15.439921; this version posted April 18, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Physiological Substrates and Ontogeny-Specific Expression of the Ubiquitin Ligases 2 MARCH1 and MARCH8 3 4 Patrick Schriek1, Haiyin Liu1, Alan C. Ching1, Pauline Huang1, Nishma Gupta1, Kayla R. 5 Wilson1, MinHsuang Tsai1, Yuting Yan2, Christophe F. Macri1, Laura F. Dagley3,4, Giuseppe 6 Infusini3,4, Andrew I. Webb3,4, Hamish McWilliam1,2, Satoshi Ishido5, Justine D. Mintern1 and 7 Jose A. Villadangos1,2 8 9 1Department of Biochemistry and Pharmacology, Bio21 Molecular Science and Biotechnology 10 Institute, The University of Melbourne, Parkville, VIC 3010, Australia. 11 2Department of Microbiology and Immunology, Peter Doherty Institute for Infection and 12 Immunity, The University of Melbourne, Parkville, VIC 3010, Australia. 13 3Advanced Technology and Biology Division, The Walter and Eliza Hall Institute of Medical 14 Research, Parkville, VIC 3052, Australia. 15 4Department of Medical Biology, University of Melbourne, Parkville, VIC 3010, Australia. 16 5Department of Microbiology, Hyogo College of Medicine, 1-1 Mukogawa-cho, Nishinomiya 17 17 663-8501, Japan 18 19 20 21 Correspondence to Justine D. Mintern ([email protected]) or 22 Jose A. Villadangos ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.04.15.439921; this version posted April 18, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Tetratricopeptide Repeat Domain 7A Is a Nuclear Factor That Modulates
El-Daher et al. Cell Discovery (2018) 4:61 Cell Discovery DOI 10.1038/s41421-018-0061-y www.nature.com/celldisc ARTICLE Open Access Tetratricopeptide repeat domain 7A is a nuclear factor that modulates transcription and chromatin structure Marie-Therese El-Daher1,2,NicolasCagnard2,3,4, Marine Gil1, Marie Chansel Da Cruz2,5, Claire Leveau1,2, Fernando Sepulveda 1,2, Mohammed Zarhrate6, Frédéric Tores3,PatriciaLegoix7, Sylvain Baulande 7, Jean Pierre de Villartay 2,5, Geneviève Almouzni8,9, Jean-Pierre Quivy8,9, Alain Fischer2,10,11,12 and GenevièvedeSaintBasile1,2,13 Abstract A loss-of-function mutation in tetratricopeptide repeat domain 7A (TTC7A) is a recently identified cause of human intestinal and immune disorders. However, clues to related underlying molecular dysfunctions remain elusive. It is now shown based on the study of TTC7A-deficient and wild-type cells that TTC7A is an essential nuclear protein. It binds to chromatin, preferentially at actively transcribed regions. Its depletion results in broad range of epigenomic changes at proximal and distal transcriptional regulatory elements and in altered control of the transcriptional program. Loss of WT_TTC7A induces general decrease in chromatin compaction, unbalanced cellular distribution of histones, higher nucleosome accessibility to nuclease digestion along with genome instability, and reduced cell viability. Our observations characterize for the first time unreported functions for TTC7A in the nucleus that exert a critical role in chromatin organization and gene regulation to safeguard healthy immune and intestinal status. 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; Introduction protein is critical for fine-tuning of the balance between The analysis of rare inherited diseases helps to identify cells’ proliferation, differentiation, and survival. -

De Novo and Rare Mutations in the HSPA1L Heat
Takahashi et al. Genome Medicine (2017) 9:8 DOI 10.1186/s13073-016-0394-9 RESEARCH Open Access De novo and rare mutations in the HSPA1L heat shock gene associated with inflammatory bowel disease Shinichi Takahashi1,2†, Gaia Andreoletti3†, Rui Chen1, Yoichi Munehira4,5, Akshay Batra6, Nadeem A. Afzal6, R. Mark Beattie6, Jonathan A. Bernstein7, Sarah Ennis3* and Michael Snyder1* Abstract Background: Inflammatory bowel disease (IBD) is a chronic, relapsing inflammatory disease of the gastrointestinal tract which includes ulcerative colitis and Crohn's disease. Genetic risk factors for IBD are not well understood. Methods: We performed a family-based whole exome sequencing (WES) analysis on a core family (Family A) to identify potential causal mutations and then analyzed exome data from a Caucasian pediatric cohort (136 patients and 106 controls) to validate the presence of mutations in the candidate gene, heat shock 70 kDa protein 1-like (HSPA1L). Biochemical assays of the de novo and rare (minor allele frequency, MAF < 0.01) mutation variant proteins further validated the predicted deleterious effects of the identified alleles. Results: In the proband of Family A, we found a heterozygous de novo mutation (c.830C > T; p.Ser277Leu) in HSPA1L. Through analysis of WES data of 136 patients, we identified five additional rare HSPA1L mutations (p.Gly77Ser, p.Leu172del, p.Thr267Ile, p.Ala268Thr, p.Glu558Asp) in six patients. In contrast, rare HSPA1L mutations were not observed in controls, and were significantly enriched in patients (P = 0.02). Interestingly, we did not find non-synonymous rare mutations in the HSP70 isoforms HSPA1A and HSPA1B. Biochemical assays revealed that all six rare HSPA1L variant proteins showed decreased chaperone activity in vitro. -

Novel Mutations of the Tetratricopeptide Repeat Domain 7A Gene and Phenotype/Genotype Comparison
ORIGINAL RESEARCH published: 07 September 2017 doi: 10.3389/fimmu.2017.01066 Novel Mutations of the Tetratricopeptide Repeat Domain 7A Gene and Phenotype/Genotype Comparison Reyin Lien 1, Yung-Feng Lin 2, Min-Wei Lai 3, Hui-Ying Weng 4, Ren-Chin Wu 5, Tang-Her Jaing 6,7, Jing-Long Huang 6,8, Shih-Feng Tsai 2 and Wen-I Lee 6,8* 1 Division of Neonatology, Department of Pediatrics, Chang Gung Memorial Hospital, Taoyuan, Taiwan, 2 Institute of Molecular and Genomic Medicine, National Health Research Institutes, Zhunan, Taiwan, 3 Division of Gastroenterology, Department of Pediatrics, Chang Gung Memorial Hospital, Taoyuan, Taiwan, 4 VYM Genome Research Center, National Yang-Ming University, Taipei, Taiwan, 5 Department of Pathology, Chang Gung Memorial Hospital, Chang Gung University, Taoyuan, Taiwan, 6 Division of Allergy, Asthma and Rheumatology, Department of Pediatrics, Chang Gung Memorial Hospital, Taoyuan, Taiwan, 7 Division of Hematology/Oncology, Department Pediatrics, Chang Gung Memorial Hospital, Taoyuan, Taiwan, 8 Primary Immunodeficiency Care and Research (PICAR) Institute, Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Taoyuan, Taiwan The gastrointestinal tract contains the largest lymphoid organ to react with pathogenic microorganisms and suppress excess inflammation. Patients with primary immunodefi- Edited by: ciency diseases (PIDs) can suffer from refractory diarrhea. In this study, we present two Isabelle Meyts, KU Leuven, Belgium siblings who began to suffer from refractory diarrhea with a poor response to aggressive Reviewed by: antibiotic and immunosuppressive treatment after surgical release of neonatal intestinal Luigi Daniele Notarangelo, obstruction. Their lymphocyte proliferation was low, but superoxide production and Harvard Medical School, IL-10 signaling were normal. -

Protein Tyrosine Phosphorylation in Haematopoietic Cancers and the Functional Significance of Phospho- Lyn SH2 Domain
Protein Tyrosine Phosphorylation in Haematopoietic Cancers and the Functional Significance of Phospho- Lyn SH2 Domain By Lily Li Jin A thesis submitted in conformity with the requirements for the degree of Ph.D. in Molecular Genetics, Graduate Department of Molecular Genetics, in the University of Toronto © Copyright by Lily Li Jin (2015) Protein Tyrosine Phosphorylation in Haematopoietic Cancers and the Functional Significance of Phospho-Lyn SH2 Domain Lily Li Jin 2015 Ph.D. in Molecular Genetics Graduate Department of Molecular Genetics University of Toronto Abstract Protein-tyrosine phosphorylation (pY) is a minor but important protein post-translational modification that modulates a wide range of cellular functions and is involved in cancer. Dysregulation of tyrosine kinases (TKs) and protein-tyrosine phosphatases (PTPs) have been observed in multiple myeloma (MM) and acute myeloid leukemia (AML) and is a subject of study. Using recently developed mass spectrometry-based proteomics techniques, quantitative PTP expression and cellular pY profiles were generated for MM cell lines and mouse xenograft tumors, as well as primary AML samples. Integrated comprehensive analyses on these data implicated a subset of TKs and PTPs in MM and AML, with valuable insights gained on the dynamic regulation of pY in biological systems. In particular, I propose a model that describes the cellular pY state as a functional output of the total activated TKs and PTPs in the cell. My results show that the global pY profile in the cancer models is quantitatively related to the cellular levels of activated TKs and PTPs. Furthermore, the identity of the implicated TK/PTPs is system- ii dependent, demonstrating context-dependent regulation of pY. -

A CASE of TTC7A DEFICIENCY Presented by Dr Bella Shadur Case
APID2016 Case Summary: A CASE OF TTC7A DEFICIENCY Presented by Dr Bella Shadur Case summary 4/12 male Non-consanguineous, Anglo-Saxon heritage Presented with severe failure to thrive and diarrhoea o Initially worked up for cow’s milk protein intolerance o No improvement with various HA formulas Colonoscopy: severe inflammation of the entire colon Referred to immunology: could this be IPEX or IL10/IL10R deficiency? Baseline immune work-up demonstrated: o IgG 0.29 (↓), IgA 0.29 (normal), IgM 0.08 (↓), IgE < 5 o Normal lymphocyte subsets o T cell phenotype: normal proliferation to PHA and anti-CD3, STAT5p, TRECs, naïve T cells o Neutrophils: normal NBT and DHR o IPEX: normal FoxP3, CD25 staining, endocrine autoantibodies NAD o HLH: ferritin 50, SAP, XIAP, perforin, NK cell degranulation normal Enrolled in NEOPICS study (Inter-National Early Onset Paediatric IBD Cohort Study) o WES: compound heterozygous for two novel mutations in the TTC7A gene o Segregated in parents (both of whom asymptomatic) o Both predicted to be pathogenic, although one required confirmation via the creation of a DNA ladder Patient stabilised on subcutaneous immunoglobulin and total parenteral nutrition Ongoing monitoring of immune parameters, particularly T cell function For consideration of bone marrow transplant Learning points TTC7A deficiency = Tetratricopeptide repeat domain 7A Located on chromosome 2p21, autosomal recessive Deficiency leads to severe pathology of the gut and the immune system o Gut disease can range from inflammatory bowel disease -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ......................................................