Single-Stranded RNA Viruses of Plants Reminiscent of Early RNA Endosymbionts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Comparison of Plant‐Adapted Rhabdovirus Protein Localization and Interactions
University of Kentucky UKnowledge University of Kentucky Doctoral Dissertations Graduate School 2011 COMPARISON OF PLANT‐ADAPTED RHABDOVIRUS PROTEIN LOCALIZATION AND INTERACTIONS Kathleen Marie Martin University of Kentucky, [email protected] Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Martin, Kathleen Marie, "COMPARISON OF PLANT‐ADAPTED RHABDOVIRUS PROTEIN LOCALIZATION AND INTERACTIONS" (2011). University of Kentucky Doctoral Dissertations. 172. https://uknowledge.uky.edu/gradschool_diss/172 This Dissertation is brought to you for free and open access by the Graduate School at UKnowledge. It has been accepted for inclusion in University of Kentucky Doctoral Dissertations by an authorized administrator of UKnowledge. For more information, please contact [email protected]. ABSTRACT OF DISSERTATION Kathleen Marie Martin The Graduate School University of Kentucky 2011 COMPARISON OF PLANT‐ADAPTED RHABDOVIRUS PROTEIN LOCALIZATION AND INTERACTIONS ABSTRACT OF DISSERTATION A dissertation submitted in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the College of Agriculture at the University of Kentucky By Kathleen Marie Martin Lexington, Kentucky Director: Dr. Michael M Goodin, Associate Professor of Plant Pathology Lexington, Kentucky 2011 Copyright © Kathleen Marie Martin 2011 ABSTRACT OF DISSERTATION COMPARISON OF PLANT‐ADAPTED RHABDOVIRUS PROTEIN LOCALIZATION AND INTERACTIONS Sonchus yellow net virus (SYNV), Potato yellow dwarf virus (PYDV) and Lettuce Necrotic yellows virus (LNYV) are members of the Rhabdoviridae family that infect plants. SYNV and PYDV are Nucleorhabdoviruses that replicate in the nuclei of infected cells and LNYV is a Cytorhabdovirus that replicates in the cytoplasm. LNYV and SYNV share a similar genome organization with a gene order of Nucleoprotein (N), Phosphoprotein (P), putative movement protein (Mv), Matrix protein (M), Glycoprotein (G) and Polymerase protein (L). -

Diversity of Plant Virus Movement Proteins: What Do They Have in Common?
processes Review Diversity of Plant Virus Movement Proteins: What Do They Have in Common? Yuri L. Dorokhov 1,2,* , Ekaterina V. Sheshukova 1, Tatiana E. Byalik 3 and Tatiana V. Komarova 1,2 1 Vavilov Institute of General Genetics Russian Academy of Sciences, 119991 Moscow, Russia; [email protected] (E.V.S.); [email protected] (T.V.K.) 2 Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia 3 Department of Oncology, I.M. Sechenov First Moscow State Medical University, 119991 Moscow, Russia; [email protected] * Correspondence: [email protected] Received: 11 November 2020; Accepted: 24 November 2020; Published: 26 November 2020 Abstract: The modern view of the mechanism of intercellular movement of viruses is based largely on data from the study of the tobacco mosaic virus (TMV) 30-kDa movement protein (MP). The discovered properties and abilities of TMV MP, namely, (a) in vitro binding of single-stranded RNA in a non-sequence-specific manner, (b) participation in the intracellular trafficking of genomic RNA to the plasmodesmata (Pd), and (c) localization in Pd and enhancement of Pd permeability, have been used as a reference in the search and analysis of candidate proteins from other plant viruses. Nevertheless, although almost four decades have passed since the introduction of the term “movement protein” into scientific circulation, the mechanism underlying its function remains unclear. It is unclear why, despite the absence of homology, different MPs are able to functionally replace each other in trans-complementation tests. Here, we consider the complexity and contradictions of the approaches for assessment of the ability of plant viral proteins to perform their movement function. -

Corona and Other Virus: Their Useful and Harmful Aspects
Current Trends on Biotechnology & Microbiology DOI: ISSN: 2641-6875 10.32474/CTBM.2020.02.000129Review Article Corona and Other Virus: Their Useful and Harmful Aspects Birhanu Gizaw* Microbial Biodiversity Directorate, Ethiopian Biodiversity Institute, Ethiopia *Corresponding author: Birhanu Gizaw, Microbial Biodiversity Directorate, Ethiopian Biodiversity Institute, P.O. Box 30726, Addis Ababa, Ethiopia Received: June 15, 2020 Published: September 22, 2020 Abstract How the single virus is forceful and shakes the world is eyewitness during this contemporary COVID 19 pandemic time. People primarily think of viruses such as HIV, Ebola, Zika, Influenza, Tobacco mosaic virus or whatever new outbreak like SARS, Corona are Understandingall viruses worst the and microbial non-beneficial. world isHowever, very critical not all and viruses crucial are thing detrimental that they and are influential driving force to human, and governing animal and the plantphysical health. world In fact, some viruses have beneficial properties for their hosts in a symbiotic relationship and scientific research in many disciplines. industry,and biosphere agriculture, at all. Thehealth virus and and environment other microbial are the life application those of bacteria, of microbes fungi, andprion, their viroid, products virion are are too requiring high for great human attention being and researchenvironment. to enhance Without their microbes utilization all lifefrom would majority be cease of useful on earth.aspects However, of microbial some genetic microbes resource. are very The dangerous secret behind like of Corona every virus, HIV, Ebola, Mycobacterium and others that destroy human life, but majority of microorganisms are too useful to promote virusdevelopment. in respect Through with health, building environment, and strengthening agriculture microbial and biotechnological culture collection application centers during and through this Covid19 strong pandemic conservation time strategy, to raise awarenessit is possible about to exploit virus atmore all. -

Small Hydrophobic Viral Proteins Involved in Intercellular Movement of Diverse Plant Virus Genomes Sergey Y
AIMS Microbiology, 6(3): 305–329. DOI: 10.3934/microbiol.2020019 Received: 23 July 2020 Accepted: 13 September 2020 Published: 21 September 2020 http://www.aimspress.com/journal/microbiology Review Small hydrophobic viral proteins involved in intercellular movement of diverse plant virus genomes Sergey Y. Morozov1,2,* and Andrey G. Solovyev1,2,3 1 A. N. Belozersky Institute of Physico-Chemical Biology, Moscow State University, Moscow, Russia 2 Department of Virology, Biological Faculty, Moscow State University, Moscow, Russia 3 Institute of Molecular Medicine, Sechenov First Moscow State Medical University, Moscow, Russia * Correspondence: E-mail: [email protected]; Tel: +74959393198. Abstract: Most plant viruses code for movement proteins (MPs) targeting plasmodesmata to enable cell-to-cell and systemic spread in infected plants. Small membrane-embedded MPs have been first identified in two viral transport gene modules, triple gene block (TGB) coding for an RNA-binding helicase TGB1 and two small hydrophobic proteins TGB2 and TGB3 and double gene block (DGB) encoding two small polypeptides representing an RNA-binding protein and a membrane protein. These findings indicated that movement gene modules composed of two or more cistrons may encode the nucleic acid-binding protein and at least one membrane-bound movement protein. The same rule was revealed for small DNA-containing plant viruses, namely, viruses belonging to genus Mastrevirus (family Geminiviridae) and the family Nanoviridae. In multi-component transport modules the nucleic acid-binding MP can be viral capsid protein(s), as in RNA-containing viruses of the families Closteroviridae and Potyviridae. However, membrane proteins are always found among MPs of these multicomponent viral transport systems. -

Plant-Based Vaccines: the Way Ahead?
viruses Review Plant-Based Vaccines: The Way Ahead? Zacharie LeBlanc 1,*, Peter Waterhouse 1,2 and Julia Bally 1,* 1 Centre for Agriculture and the Bioeconomy, Queensland University of Technology (QUT), Brisbane, QLD 4000, Australia; [email protected] 2 ARC Centre of Excellence for Plant Success in Nature and Agriculture, Queensland University of Technology (QUT), Brisbane, QLD 4000, Australia * Correspondence: [email protected] (Z.L.); [email protected] (J.B.) Abstract: Severe virus outbreaks are occurring more often and spreading faster and further than ever. Preparedness plans based on lessons learned from past epidemics can guide behavioral and pharmacological interventions to contain and treat emergent diseases. Although conventional bi- ologics production systems can meet the pharmaceutical needs of a community at homeostasis, the COVID-19 pandemic has created an abrupt rise in demand for vaccines and therapeutics that highlight the gaps in this supply chain’s ability to quickly develop and produce biologics in emer- gency situations given a short lead time. Considering the projected requirements for COVID-19 vaccines and the necessity for expedited large scale manufacture the capabilities of current biologics production systems should be surveyed to determine their applicability to pandemic preparedness. Plant-based biologics production systems have progressed to a state of commercial viability in the past 30 years with the capacity for production of complex, glycosylated, “mammalian compatible” molecules in a system with comparatively low production costs, high scalability, and production flexibility. Continued research drives the expansion of plant virus-based tools for harnessing the full production capacity from the plant biomass in transient systems. -

The Evolution of Genome Compression and Genomic Novelty in RNA Viruses
Downloaded from genome.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Letter The evolution of genome compression and genomic novelty in RNA viruses Robert Belshaw,1,3 Oliver G. Pybus,1 and Andrew Rambaut2 1Department of Zoology, University of Oxford, Oxford OX1 3PS, United Kingdom; 2Institute of Evolutionary Biology, University of Edinburgh, Edinburgh EH9 3JT, United Kingdom The genomes of RNA viruses are characterized by their extremely small size and extremely high mutation rates (typically 10 kb and 10−4/base/replication cycle, respectively), traits that are thought to be causally linked. One aspect of their small size is the genome compression caused by the use of overlapping genes (where some nucleotides code for two genes). Using a comparative analysis of all known RNA viral species, we show that viruses with larger genomes tend to have less gene overlap. We provide a numerical model to show how a high mutation rate could lead to gene overlap, and we discuss the factors that might explain the observed relationship between gene overlap and genome size. We also propose a model for the evolution of gene overlap based on the co-opting of previously unused ORFs, which gives rise to two types of overlap: (1) the creation of novel genes inside older genes, predominantly via +1 frameshifts, and (2) the incremental increase in overlap between originally contiguous genes, with no frameshift preference. Both types of overlap are viewed as the creation of genomic novelty under pressure for genome compression. Simulations based on our model generate the empirical size distributions of overlaps and explain the observed frameshift preferences. -

Plant Virus RNA Replication
eLS Plant Virus RNA Replication Alberto Carbonell*, Juan Antonio García, Carmen Simón-Mateo and Carmen Hernández *Corresponding author: Alberto Carbonell ([email protected]) A22338 Author Names and Affiliations Alberto Carbonell, Instituto de Biología Molecular y Celular de Plantas (CSIC-UPV), Campus UPV, Valencia, Spain Juan Antonio García, Centro Nacional de Biotecnología (CSIC), Madrid, Spain Carmen Simón-Mateo, Centro Nacional de Biotecnología (CSIC), Madrid, Spain Carmen Hernández, Instituto de Biología Molecular y Celular de Plantas (CSIC-UPV), Campus UPV, Valencia, Spain *Advanced article Article Contents • Introduction • Replication cycles and sites of replication of plant RNA viruses • Structure and dynamics of viral replication complexes • Viral proteins involved in plant virus RNA replication • Host proteins involved in plant virus RNA replication • Functions of viral RNA in genome replication • Concluding remarks Abstract Plant RNA viruses are obligate intracellular parasites with single-stranded (ss) or double- stranded RNA genome(s) generally encapsidated but rarely enveloped. For viruses with ssRNA genomes, the polarity of the infectious RNA (positive or negative) and the presence of one or more genomic RNA segments are the features that mostly determine the molecular mechanisms governing the replication process. RNA viruses cannot penetrate plant cell walls unaided, and must enter the cellular cytoplasm through mechanically-induced wounds or assisted by a 1 biological vector. After desencapsidation, their genome remains in the cytoplasm where it is translated, replicated, and encapsidated in a coupled manner. Replication occurs in large viral replication complexes (VRCs), tethered to modified membranes of cellular organelles and composed by the viral RNA templates and by viral and host proteins. -
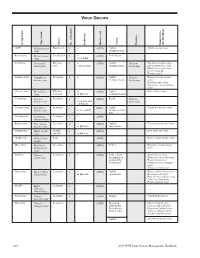
Virus Groups 2 1 Group Name Group Strain Type Shape No
VIRUS GROUPS 2 1 Group Name Group Strain Type Shape No. of Particles Inclusions Nucleic Acid Vector Features Members in this Handbook AMV Alfalfa mosaic Baciliform 4 + ssRNA Aphid Alfalfa mosaic virus virus (nonpersistent) Bromovirus Brome mosiac Isosahedral 3 + ssRNA Some beetle virus crystalline Carlavirus Carnation Flexous 1 + ssRNA Aphid Narrow Blackberry calico virus latent virus rod various types (nonpersistent) host range Lily symptomless virus Blueberry scorch virus Potato virus M Potato virus S Caulimovirus Cauliflower Isometric 1 + dsDNA Aphid Narrow Blueberry red ringspot mosaic virus (nonpersistent) host range virus Dahlia nosaic virus Strawberry vein-banding virus Closterovirus Beet yellows Filamen- 1 + ssRNA Aphid Beet yellows virus virus tous rod in phloem (semipersistent) Comovirus Cowpea Isometric 2 + ssRNA Beetle Narrow mosaic virus vacuolate and host range crystalloid Cucumovirus Cucumber Isometric 3 + sRNA Aphid Cucumber mosaic virus mosaic virus in mesophyll (nonpersistent), Sap Dianthovirus Carnation Isometric 2 + ssRNA ringspot virus Enamovirus Pea enation Isometric 2 + ssRNA Aphid Pea enation mosaic virus mosaic virus in phloem (persistent) Geminivirus Maize streak Double 1 + ssDNA Beet curly top virus virus particle in phloem Hordeivirus Barley stripe Rod 3 ssRNA Barley stripe mosaic virus mosaic Ideaovirus Raspberry Isometric 3 ssRNA Pollen Raspberry bushy dwarf bushy dwarf virus virus Ilarvirus Tobacco Isometric 3 + ssRNA Pollen, Seed Apple mosaic virus streak virus (transmission Blueberry shock ilarvirus mediated -

Principles and Applications in Plant Virology
plants Review Loop Mediated Isothermal Amplification: Principles and Applications in Plant Virology 1, , 2, 3, 1, Stefano Panno * y, Slavica Mati´c y, Antonio Tiberini y, Andrea Giovanni Caruso y , Patrizia Bella 1, Livio Torta 1 , Raffaele Stassi 1 and Salvatore Davino 1,4,* 1 Department of Agricultural, Food and Forest Sciences, University of Palermo, 90128 Palermo, Italy; [email protected] (A.G.C.); [email protected] (P.B.); [email protected] (L.T.); [email protected] (R.S.) 2 Department of Agricultural, Forestry and Food Sciences, University of Turin, 10095 Turin, Italy; [email protected] 3 Council for Agricultural Research and Economics, Research Center for Plant Protection and Certification, 00156 Rome, Italy; [email protected] 4 Institute for Sustainable Plant Protection, National Research Council (IPSP-CNR), 10135 Turin, Italy * Correspondence: [email protected] (S.P.); [email protected] (S.D.); Tel.: +39-0912-389-6049 (S.P. & S.D.) These authors contributed equally to this work. y Received: 6 March 2020; Accepted: 2 April 2020; Published: 6 April 2020 Abstract: In the last decades, the evolution of molecular diagnosis methods has generated different advanced tools, like loop-mediated isothermal amplification (LAMP). Currently, it is a well-established technique, applied in different fields, such as the medicine, agriculture, and food industries, owing to its simplicity, specificity, rapidity, and low-cost efforts. LAMP is a nucleic acid amplification under isothermal conditions, which is highly compatible with point-of-care (POC) analysis and has the potential to improve the diagnosis in plant protection. The great advantages of LAMP have led to several upgrades in order to implement the technique. -

Virus Detection in Banana a Laboratory Manual
Virus Detection in Banana A Laboratory Manual www.iita.org Virus Detection in Banana A Laboratory Manual Edited and Compiled by P Lava Kumar Virology & Molecular Diagnostics Unit International Institute of Tropical Agriculture PMB 5320, Ibadan, Nigeria www.iita.org About IITA The International Institute of Tropical Agriculture (IITA) is an international non-profit R4D organization founded in 1967 as a research institute with a mandate to develop sustainable food production systems in tropical Africa. It became the first African link in the worldwide network of agricultural research centers supported by the Consultative Group on International Agricultural Research (CGIAR) formed in 1971. Mandate crops of IITA include banana & plantain, cassava, cowpea, maize, soybean and yam. IITA operates throughout sub-Saharan Africa and has headquarters in Ibadan, Nigeria. IITA’s mission is to enhance food security and improve livelihoods in Africa through research for development (R4D), a process where science is employed to identify problems and to create development solutions which result in local production, wealth creation, and the reduction of risk. IITA works with the partners within Africa and beyond. For further details about the organization visit: www.iita.org. Headquarters: International mailing address: IITA, PMB 5320, Ibadan, Oyo State, Nigeria IITA, Carolyn House, Tel: +234 2 7517472, (0)8039784000 26 Dingwall Road, Croydon Fax: INMARSAT: 873761798636 CR9 3EE, England E-mail: [email protected] United Kingdom Copyright © 2010 IITA This manual is a semi-formal publication. IITA holds the copyright to its publications but encourages duplication of these materials for noncommercial purposes. Proper citation is requested and prohibits modification of these materials. -
Protein Structure-Guided Hidden Markov Models (Hmms) As a Powerful Method in the Detection of Ancestral Endogenous Viral Elements
viruses Article Protein Structure-Guided Hidden Markov Models (HMMs) as A Powerful Method in the Detection of Ancestral Endogenous Viral Elements Heleri Kirsip 1,* and Aare Abroi 2,* 1 Department of Bioinformatics, University of Tartu, Tartu, 51010, Riia 23, Estonia 2 Institute of Technology, University of Tartu, Tartu, 50411, Nooruse 1, Estonia * Correspondence: [email protected] (H.K.); [email protected] (A.A.) Received: 29 January 2019; Accepted: 27 March 2019; Published: 2 April 2019 Abstract: It has been believed for a long time that the transfer and fixation of genetic material from RNA viruses to eukaryote genomes is very unlikely. However, during the last decade, there have been several cases in which “virus-to-host” gene transfer from various viral families into various eukaryotic phyla have been described. These transfers have been identified by sequence similarity, which may disappear very quickly, especially in the case of RNA viruses. However, compared to sequences, protein structure is known to be more conserved. Applying protein structure-guided protein domain-specific Hidden Markov Models, we detected homologues of the Virgaviridae capsid protein in Schizophora flies. Further data analysis supported “virus-to-host” transfer into Schizophora ancestors as a single transfer event. This transfer was not identifiable by BLAST or by other methods we applied. Our data show that structure-guided Hidden Markov Models should be used to detect ancestral virus-to-host transfers. Keywords: endogenous viral elements; bioinformatics; horizontal gene transfer; virus-to-host gene transfer; HMM; tobacco mosaic virus; Drosophila; capsid protein 1. Introduction Viruses are one of the most abundant and prevalent biological entities on Earth and thus are an important and integral part of the biosphere (for viral importance in environments, see [1–4]). -
Plant Viruses: from Targets to Tools for CRISPR
viruses Review Plant Viruses: From Targets to Tools for CRISPR Carla M. R. Varanda 1,* , Maria do Rosário Félix 2, Maria Doroteia Campos 1 , Mariana Patanita 1 and Patrick Materatski 1,* 1 MED—Mediterranean Institute for Agriculture, Environment and Development, Instituto de Investigação e Formação Avançada, Universidade de Évora, Pólo da Mitra, Ap. 94, 7006-554 Évora, Portugal; [email protected] (M.D.C.); [email protected] (M.P.) 2 MED—Mediterranean Institute for Agriculture, Environment and Development & Departamento de Fitotecnia, Escola de Ciências e Tecnologia, Universidade de Évora, Pólo da Mitra, Ap. 94, 7006-554 Évora, Portugal; [email protected] * Correspondence: [email protected] (C.M.R.V.); [email protected] (P.M.) Abstract: Plant viruses cause devastating diseases in many agriculture systems, being a serious threat for the provision of adequate nourishment to a continuous growing population. At the present, there are no chemical products that directly target the viruses, and their control rely mainly on preventive sanitary measures to reduce viral infections that, although important, have proved to be far from enough. The current most effective and sustainable solution is the use of virus-resistant varieties, but which require too much work and time to obtain. In the recent years, the versatile gene editing technology known as CRISPR/Cas has simplified the engineering of crops and has successfully been used for the development of viral resistant plants. CRISPR stands for ‘clustered regularly interspaced short palindromic repeats’ and CRISPR-associated (Cas) proteins, and is based on a natural adaptive immune system that most archaeal and some bacterial species present to defend themselves against invading bacteriophages.