The Mechanism of Integration Preference to Heterochromatin of Yeast Retrotransposon Ty5 Weiwu Xie Iowa State University
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

"Evolutionary Emergence of Genes Through Retrotransposition"
Evolutionary Emergence of Advanced article Genes Through Article Contents . Introduction Retrotransposition . Gene Alteration Following Retrotransposon Insertion . Retrotransposon Recruitment by Host Genome . Retrotransposon-mediated Gene Duplication Richard Cordaux, University of Poitiers, Poitiers, France . Conclusion Mark A Batzer, Department of Biological Sciences, Louisiana State University, Baton Rouge, doi: 10.1002/9780470015902.a0020783 Louisiana, USA Variation in the number of genes among species indicates that new genes are continuously generated over evolutionary times. Evidence is accumulating that transposable elements, including retrotransposons (which account for about 90% of all transposable elements inserted in primate genomes), are potent mediators of new gene origination. Retrotransposons have fostered genetic innovation during human and primate evolution through: (i) alteration of structure and/or expression of pre-existing genes following their insertion, (ii) recruitment (or domestication) of their coding sequence by the host genome and (iii) their ability to mediate gene duplication via ectopic recombination, sequence transduction and gene retrotransposition. Introduction genes, respectively, and de novo origination from previ- ously noncoding genomic sequence. Genome sequencing Variation in the number of genes among species indicates projects have also highlighted that new gene structures can that new genes are continuously generated over evolution- arise as a result of the activity of transposable elements ary times. Although the emergence of new genes and (TEs), which are mobile genetic units or ‘jumping genes’ functions is of central importance to the evolution of that have been bombarding the genomes of most species species, studies on the formation of genetic innovations during evolution. For example, there are over three million have only recently become possible. -
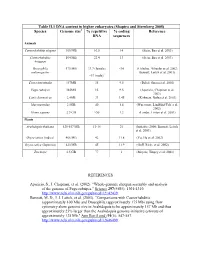
Table II.1 DNA Content in Higher Eukaryotes (Shapiro and Sternberg 2005) Species Genome Size % Repetitive DNA % Coding Sequences
Table II.1 DNA content in higher eukaryotes (Shapiro and Sternberg 2005) Species Genome size1 % repetitive % coding Reference DNA sequences Animals Caenorhabditis elegans 100 MB 16.5 14 (Stein, Bao et al. 2003) Caenorhabditis 104 MB 22.4 13 (Stein, Bao et al. 2003) briggsae Drosophila 175 MB 33.7 (female) <10 (Celniker, Wheeler et al. 2002; melanogaster Bennett, Leitch et al. 2003) ~57 (male)2 Ciona intestinalis 157MB 35 9.5 (Dehal, Satou et al. 2002) Fugu rubripes 365MB 15 9.5 (Aparicio, Chapman et al. 2002) Canis domesticus 2.4GB 31 1.45 (Kirkness, Bafna et al. 2003) Mus musculus 2.5GB 40 1.4 (Waterston, Lindblad-Toh et al. 2002) Homo sapiens 2.9 GB >50 1.2 (Lander, Linton et al. 2001) Plants Arabidopsis thaliana 125-157 MB 13-14 21 (Initiative 2000; Bennett, Leitch et al. 2003) Oryza sativa (indica) 466 MB 42 11.8 (Yu, Hu et al. 2002) Oryza sativa (Japonica) 420 MB 45 11.9 (Goff, Ricke et al. 2002) Zea mays 2.5 GB 77 1 (Meyers, Tingey et al. 2001) REFERENCES Aparicio, S., J. Chapman, et al. (2002). "Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes." Science 297(5585): 1301-1310. http://www.ncbi.nlm.nih.gov/pubmed/12142439. Bennett, M. D., I. J. Leitch, et al. (2003). "Comparisons with Caenorhabditis (approximately 100 Mb) and Drosophila (approximately 175 Mb) using flow cytometry show genome size in Arabidopsis to be approximately 157 Mb and thus approximately 25% larger than the Arabidopsis genome initiative estimate of approximately 125 Mb." Ann Bot (Lond) 91(5): 547-557. -

B2 and ALU Retrotransposons Are Self-Cleaving Ribozymes Whose Activity Is Enhanced by EZH2
B2 and ALU retrotransposons are self-cleaving ribozymes whose activity is enhanced by EZH2 Alfredo J. Hernandeza,1, Athanasios Zovoilisb,c,1,2, Catherine Cifuentes-Rojasb,c,1,3, Lu Hanb,c, Bojan Bujisicb,c, and Jeannie T. Leeb,c,4 aDepartment of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115; bDepartment of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114; and cDepartment of Genetics, The Blavatnik Institute, Harvard Medical School, Boston, MA 02114 Contributed by Jeannie T. Lee, November 18, 2019 (sent for review October 9, 2019; reviewed by Vivian G. Cheung and Karissa Y. Sanbonmatsu) Transposable elements make up half of the mammalian genome. B2s are present in ∼350,000 copies in the mouse genome (10) One of the most abundant is the short interspersed nuclear and give rise to 180- to 200-nucleotide (nt) polymerase III complex element (SINE). Among their million copies, B2 accounts for (POL-III) transcripts. B2 SINEs are transcriptionally active, with ∼350,000 in the mouse genome and has garnered special interest expression being highest in germ cells and in early development, because of emerging roles in epigenetic regulation. Our recent work after which they become epigenetically silenced. However, one of demonstrated that B2 RNA binds stress genes to retard transcription the most intriguing aspects of SINE biology is that these elements elongation. Although epigenetically silenced, B2s become massively become highly up-regulated when somatic cells are acutely stressed up-regulated during thermal and other types of stress. Specifically, (11–13), including during viral infection (8, 11), autoimmune pro- an interaction between B2 RNA and the Polycomb protein, EZH2, cesses such as macular degeneration (14, 15), and progression to results in cleavage of B2 RNA, release of B2 RNA from chromatin, cancer (16, 17). -

The Saccharomyces Ty5 Retrotransposon Family Is Associated with Origins of DNA Replication at the Telomeres and the Silent Mating Locus HMR SIGE Zou, DAVID A
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 920-924, January 1995 Genetics The Saccharomyces Ty5 retrotransposon family is associated with origins of DNA replication at the telomeres and the silent mating locus HMR SIGE Zou, DAVID A. WRIGHT, AND DANIEL F. VOYTAS* Department of Zoology and Genetics, Iowa State University, Ames, IA 50011 Communicated by Mary Lou Pardue, Massachusetts Institute of Technology, Cambridge, MA, October 3, 1994 ABSTRACT We have characterized the genomic organi- with tRNA genes has been well documented, and most Ty3 zation of the TyS retrotransposons among diverse strains of insertions are located within a few bases of the transcription Saccharomyces cerevisiae and the related species Saccharomyces start site of genes transcribed by RNA polymerase III (pol III) paradoxus. The S. cerevisiae strain S288C (or its derivatives) (4). The fourth retrotransposon family, Ty4, is represented by carries eight Ty5 insertions. Six of these are located near the a single insertion on chr III. This element is within 300 bp of telomeres, and five are found within 500 bp of autonomously a tRNA gene, and tRNA genes are associated with 10 of 12 Ty4 replicating sequences present in the type X subtelomeric insertions currently in the sequence data base (GenBank, repeat. The remaining two S. cerevisiae elements are adjacent release 83.0). Thus, a target bias, and in particular a preference to the silent mating locus HMR and are located within 500 bp for tRNA genes, is readily apparent from the genomic orga- of the origin of replication present in the transcriptional nization of endogenous Tyl-4 elements. -

Stable Integration of Transgenes Delivered by a Retrotransposon–Adenovirus Hybrid Vector
HUMAN GENE THERAPY 12:1417–1428 (July 20, 2001) Mary Ann Liebert, Inc. Stable Integration of Transgenes Delivered by a Retrotransposon–Adenovirus Hybrid Vector HARRIS SOIFER, 1,2 COLLIN HIGO, 1,2 HAIG H. KAZAZIAN, JR., 3 JOHN V. MORAN, 4 KOHNOSUKE MITANI, 5 and NORIYUKI KASAHARA 1,2,6 ABSTRACT Helper-dependent adenoviruses show great promise as gene delivery vectors. However, because they do not integrate into the host chromosome, transgene expression cannot be maintained indefinitely. To overcome these limitations, we have inserted an L1 retrotransposon/transgene element into a helper-dependent adeno- virus to create a novel chimeric gene delivery vector. Efficient adenovirus-mediated delivery of the L1 ele- ment into cultured human cells results in subsequent retrotransposition and stable integration of the trans- gene. L1 retrotransposition frequency was found to correlate with increasing multiplicity of infection by the chimeric vector, and further retrotransposition from newly integrated elements was not observed on pro- longed culture. Therefore, this vector, which utilizes components of low immunogenic potential, represents a novel two-stage gene delivery system capable of achieving high titers via the initial helper-dependent adeno- virus stage and permanent transgene integration via the retrotransposition stage. OVERVIEW SUMMARY ing to achieve efficient gene transfer, a variety of viruses have been adapted for use as gene delivery vectors, including retro- A retrotransposition-competent LINE-1 element (RC-L1) virus, adenovirus, and adeno-associated virus. Of these, ade- was inserted into a helper-dependent adenovirus (HDAd) to noviral vectors are capable of achieving high titers and have generate a hybrid adenovirus–retrotransposon vector. -

The Enigma of Y Chromosome Degeneration: T', a Novel Retrotransposon Is Preferentially Located on the Neey Chromosome of Drosophila Miranda
Copyright 0 1997 by the Genetics Society of America The Enigma of Y Chromosome Degeneration: T', a Novel Retrotransposon is Preferentially Located on the NeeY Chromosome of Drosophila miranda Manfred Steinemann and Sigrid Steinemann Institut fur Genetik, Heinrich-Heine-Universitat Dusseldorf; 0-40225 Dusseldorf, Germany Manuscript received July 16, 1996 Accepted for publication October 17, 1996 ABSTRACT We have cloned a novel transposable element from the nepY chromosome of Drosophila miranda. The size of the element, designated asTRAM, is 3.452 bp, including on both sides long terminal direct repeats (LTRs)of 372 bp, respectively. The element is flanked by a 5-bp target site duplication, ATATG. The putative primer binding site (PBS) for minus-strand priming is complementary to 18 nucleotides of the 3"end of tRNATv. Data base screens for DNA sequence identities were negative, apart from the sequence motif of the PBS. The deduced amino acid sequence from the large ORF does not' reveal identities described for other transposons. In situ hybridizations with TRAM subclones show a biased distribution in the genome, with a massive accumulation of TRAM in the neo-Y chromosome, while the former homologue, the XZchromosomeis devoid of TRAMsites. The enriched occurrence of the TRAM element at the evolving neo-Y chromosome of D.miranda adds compelling evidencein favor of the view that Y chromosome degeneration is driven by the accumulation of transposable elements. CHROMOSOME degeneration (MULLER1918, polytene chromosome squashes, the male X chromo- Y 1932) is a process that involves structural changes some in Drosophila can be distinguished by the pres- in chromosome architecture and expansionof genetic ence of an isoform of histone H4 acetylated at lysine 16, inertness along theY chromosome (6CHARLESWORTH H4.Ac16. -

Like Retrotransposon (Transposable Dement/Interspedflc Hybridization/Zea/Tnpsacum/Waxy Alde) MICHAEL D
Proc. Nati. Acad. Sci. USA Vol. 91, pp. 11674-11678, November 1994 Evolution Molecular evolution of magellan, a maize Ty3/gypsy- like retrotransposon (transposable dement/interspedflc hybridization/Zea/Tnpsacum/waxy alde) MICHAEL D. PURUGGANAN*t AND SUSAN R. WESSLER*t§ Departments of *Botany and *Genetics, University of Georgia, Athens, GA 30602 Communicated by Wyatt W. Anderson, July 28, 1994 ABSTRACT The magelan transposable element is respon- Understanding the evolutionary biology of retrotrans- sible for a spontaneous 5.7-kb insertion in the maize wx-M posons requires data on the extent and patterns of molecular allele. This element has the sequence and structural charac- variation within element families. In this report, we describe teristics of a Ty3/gypsy-like retrotransposon. The maeln a low-copy-number retrotransposon whose presence within element is present in all Zea species and Tripsacum andersonu; the phylogenetically well-defined genus Zea makes it an It is absent, however, in the genomes of all other Tripsacum excellent subject for the study of retrotransposon evolution. species analyzed. The genetic distances between magellan ele- This Ty3/gypsy-like retrotransposon, which we have named ments suggest that this retrotransposon is evolving faster than magellan, is responsible for a 5.7-kb insertion in a sponta- other Zea nuclear loci. The phylogeny of magelan within Zea neous mutant allele of the maize waxy (wx) locus. The and T. andersonf also reveals a pattern of interspecies trans- magellan element is also found in Tripsacum andersonii but fers, resulting in the movement of mageflan subfmles be- is absent in all other Tripsacum species tested. -

The Repetitive Landscape of the 5100 Mbp Barley Genome Thomas Wicker1*, Alan H
Wicker et al. Mobile DNA (2017) 8:22 DOI 10.1186/s13100-017-0102-3 RESEARCH Open Access The repetitive landscape of the 5100 Mbp barley genome Thomas Wicker1*, Alan H. Schulman2,3, Jaakko Tanskanen2,3, Manuel Spannagl4, Sven Twardziok4, Martin Mascher5,6, Nathan M. Springer7, Qing Li7,8, Robbie Waugh9,10, Chengdao Li11,12, Guoping Zhang13, Nils Stein5, Klaus F. X. Mayer4,14 and Heidrun Gundlach4 Abstract Background: While transposable elements (TEs) comprise the bulk of plant genomic DNA, how they contribute to genome structure and organization is still poorly understood. Especially in large genomes where TEs make the majority of genomic DNA, it is still unclear whether TEs target specific chromosomal regions or whether they simply accumulate where they are best tolerated. Results: Here, we present an analysis of the repetitive fraction of the 5100 Mb barley genome, the largest angiosperm genome to have a near-complete sequence assembly. Genes make only about 2% of the genome, while over 80% is derived from TEs. The TE fraction is composed of at least 350 different families. However, 50% of the genome is comprised of only 15 high-copy TE families, while all other TE families are present in moderate or low copy numbers. We found that the barley genome is highly compartmentalized with different types of TEs occupying different chromosomal “niches”, such as distal, interstitial, or proximal regions of chromosome arms. Furthermore, gene space represents its own distinct genomic compartment that is enriched in small non-autonomous DNA transposons, suggesting that these TEs specifically target promoters and downstream regions. -

Downloads/Repeatmaskedgenomes
Kojima Mobile DNA (2018) 9:2 DOI 10.1186/s13100-017-0107-y REVIEW Open Access Human transposable elements in Repbase: genomic footprints from fish to humans Kenji K. Kojima1,2 Abstract Repbase is a comprehensive database of eukaryotic transposable elements (TEs) and repeat sequences, containing over 1300 human repeat sequences. Recent analyses of these repeat sequences have accumulated evidences for their contribution to human evolution through becoming functional elements, such as protein-coding regions or binding sites of transcriptional regulators. However, resolving the origins of repeat sequences is a challenge, due to their age, divergence, and degradation. Ancient repeats have been continuously classified as TEs by finding similar TEs from other organisms. Here, the most comprehensive picture of human repeat sequences is presented. The human genome contains traces of 10 clades (L1, CR1, L2, Crack, RTE, RTEX, R4, Vingi, Tx1 and Penelope) of non-long terminal repeat (non-LTR) retrotransposons (long interspersed elements, LINEs), 3 types (SINE1/7SL, SINE2/tRNA, and SINE3/5S) of short interspersed elements (SINEs), 1 composite retrotransposon (SVA) family, 5 classes (ERV1, ERV2, ERV3, Gypsy and DIRS) of LTR retrotransposons, and 12 superfamilies (Crypton, Ginger1, Harbinger, hAT, Helitron, Kolobok, Mariner, Merlin, MuDR, P, piggyBac and Transib) of DNA transposons. These TE footprints demonstrate an evolutionary continuum of the human genome. Keywords: Human repeat, Transposable elements, Repbase, Non-LTR retrotransposons, LTR retrotransposons, DNA transposons, SINE, Crypton, MER, UCON Background contrast, MER4 was revealed to be comprised of LTRs of Repbase and conserved noncoding elements endogenous retroviruses (ERVs) [1]. Right now, Repbase Repbase is now one of the most comprehensive data- keeps MER1 to MER136, some of which are further bases of eukaryotic transposable elements and repeats divided into several subfamilies. -

Developmental Retrotransposon Activation Primes Host Immunity for Future Viral- Clearance
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.23.263293; this version posted August 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Developmental retrotransposon activation primes host immunity for future viral- clearance Lu Wang1, Lauren Tracy1, ZZ Zhao Zhang1,2,* 1Department of Pharmacology & Cancer Biology, Duke University School of Medicine, Durham, USA. 2Regeneration Next, Duke University School of Medicine, Durham, USA. *Corresponding author: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.23.263293; this version posted August 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Transposons are thought to be largely suppressed under physiological conditions, ensuring that their mobilization is a rare event. By tracking mobilization, we show that during metamorphosis at the Drosophila pupal stage, the Gypsy retrotransposon selectively mobilizes in regenerating tissues. In the newly formed tissues, this wave of Gypsy activation primes the host’s innate immune system by inducing the production of antimicrobial peptides (AMPs). Moreover, early immune-priming functions of Gypsy are essential for combating viral invasion in adult flies: flies with Gypsy being silenced at the pupal stage are unable to clear viruses and succumb to viral infection. Our data reveal that regulated activation of transposons during animal developmental endows a long-term benefit in pathogen warfare. 2 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.23.263293; this version posted August 24, 2020. -

Arabidopsis Retrotransposon Virus-Like Particles and Their Regulation by Epigenetically Activated Small RNA
Downloaded from genome.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Research Arabidopsis retrotransposon virus-like particles and their regulation by epigenetically activated small RNA Seung Cho Lee,1,3 Evan Ernst,1,3 Benjamin Berube,2 Filipe Borges,1 Jean-Sebastien Parent,1 Paul Ledon,2 Andrea Schorn,2 and Robert A. Martienssen1,2 1Howard Hughes Medical Institute, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA; 2Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA In Arabidopsis, LTR retrotransposons are activated by mutations in the chromatin gene DECREASE in DNA METHYLATION 1 (DDM1), giving rise to 21- to 22-nt epigenetically activated siRNA (easiRNA) that depend on RNA DEPENDENT RNA POLYMERASE 6 (RDR6). We purified virus-like particles (VLPs) from ddm1 and ddm1rdr6 mutants in which genomic RNA is reverse transcribed into complementary DNA. High-throughput short-read and long-read sequencing of VLP DNA (VLP DNA-seq) revealed a comprehensive catalog of active LTR retrotransposons without the need for mapping transpo- sition, as well as independent of genomic copy number. Linear replication intermediates of the functionally intact COPIA element EVADE revealed multiple central polypurine tracts (cPPTs), a feature shared with HIV in which cPPTs promote nu- clear localization. For one member of the ATCOPIA52 subfamily (SISYPHUS), cPPT intermediates were not observed, but abundant circular DNA indicated transposon “suicide” by auto-integration within the VLP. easiRNA targeted EVADE geno- mic RNA, polysome association of GYPSY (ATHILA) subgenomic RNA, and transcription via histone H3 lysine-9 dimethy- lation. VLP DNA-seq provides a comprehensive landscape of LTR retrotransposons and their control at transcriptional, post-transcriptional, and reverse transcriptional levels. -

Factors That Affect Targeting of Ty5 Retrotransposon in Saccharomyces Cerevisiae Robert Dick Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 Factors that affect targeting of Ty5 retrotransposon in Saccharomyces cerevisiae Robert Dick Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Genetics and Genomics Commons Recommended Citation Dick, Robert, "Factors that affect targeting of Ty5 retrotransposon in Saccharomyces cerevisiae" (2007). Retrospective Theses and Dissertations. 15047. https://lib.dr.iastate.edu/rtd/15047 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Factors that affect targeting of Ty5 retrotransposon in Saccharomyces cerevisiae by Robert Dick A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Major: Interdepartmental Genetics Program of Study Committee: Daniel F. Voytas, Major Professor Linda Abrosio Diane Bassham Iowa State University Ames, Iowa 2007 Copyright Robert Dick, 2007. All rights reserved. UMI Number: 1446046 Copyright 2007 by Dick, Robert All rights reserved. ____________________________________________________________ UMI Microform 1446046 Copyright 2007 by ProQuest Information and Learning Company.