Integration Specificity of the Retrovirus-Like Transposable Element Ty5 of Saccharomyces Sige Zou Iowa State University
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Utility of Subtelomeric Fluorescent DNA Probes for Detection of Chromosome Anomalies in 425 Patients Syed M
article January/February 2003 ⅐ Vol. 5 ⅐ No. 1 Utility of subtelomeric fluorescent DNA probes for detection of chromosome anomalies in 425 patients Syed M. Jalal, PhD1, Aaron R. Harwood1, Gurbax S. Sekhon, PhD3, Cindy Pham Lorentz, MS1, Rhett P. Ketterling, MD1, Dusica Babovic-Vuksanovic, MD2, Reid G. Meyer1, Regina Ensenauer, MD2, Marvin H. Anderson, Jr1, and Virginia V. Michels, MD2 Purpose: A complete set of subtelomeric fluorescent DNA probes, except the acrocentric p-arms, was developed in 1996, was optimized in 1998, and is commercially available. These and other fluorescence in situ hybridization (FISH) probes have been used to detect anomalies of the subtelomere regions among groups of patients with idiopathic mental retardation (MR), developmental delay (DD), and/or nonspecific dysmorphic features (NDF), and individuals with multiple miscarriages (MM) who were karyotypically normal by standard G-banding techniques. Methods: A total of 425 patients were analyzed, of whom 372 had idiopathic MR/DD/NDF and 53 were involved in MM. An effort was made to select individuals for this study who were either normal karyotypically or who had subtle chromosomal anomalies that were inconclusive by banded chromosome analysis, although this was not always possible. Results: Anomalies involving the subtelomere regions were detected at a frequency of 6.8% in the MR/DD/NDF group. The cryptic or subtle anomalies are estimated to be about 3.4%. It was necessary to use M-FISH, chromosome, and locus specific FISH probes to clarify some of the abnormalities. No abnormalities were detected in the MM group. Deletion variants were present for 2qter, 7pter, and Xpter/Ypter subtelomeric regions ranging from Ͻ1 to 9.6%. -

Harnessing Single-Molecule Sequencing to Characterize the Fast-Evolving Drosophila Subtelomere Xander M
Harnessing single-molecule sequencing to characterize the fast-evolving Drosophila subtelomere Xander M. Gottfried, COL 2021 Mia T. Levine, College of Arts and Sciences Department of Biology Abstract ORF polymorphism is concentrated closer to telomere The telomere and subtelomere are repetitive sequences at the ends of chromosomes Use genome BLAST to find subtelomeric genes required for chromosome length preservation. In Drosophila, telomere and subtelomere are highly plastic; each of them varies in copy number and sequence both within and across species. In addition, there is evidence of functional crosstalk between telomere and subtelomere, suggesting that the two regions may co-evolve to maintain system fidelity. However, without characterizing the sequence of the subtelomere, we cannot investigate whether subtelomere evolution affects telomere function. This characterization has recently been made possible due to the advent of single-molecule sequencing, which can be used to assemble repetitive regions using long, 100 kilobase reads. Here, we begin to characterize the composition and variability of subtelomeric genes, focusing on exon duplications, intergenic distance variability, and functional open reading frame polymorphism. The Drosophila Subtelomere • Highly variable in copy number and sequence within species • Rapidly evolving Exon fragment duplications are more common closer across species to the telomere • Pervasive terminal Chromosome 2L Average # Exon Fragment Duplications Across Genomes 12 deletions • Functional Experimental Validation: crosstalk with 8 • PCR: primers to absent genes, primers to unorthodox telomere has break points, primers across gaps implications for • Cell biology: DNA FISH to gene sequences, IF to 4 genome integrity proteins RNAseq to dysfunctional ORFs Average # ofExon FragmentDuplications # Average 0 References: Anderson, J.A., Song, Y.S., and Langley, C.H. -
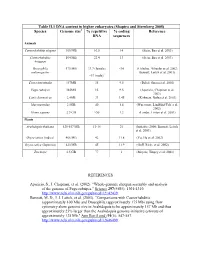
Table II.1 DNA Content in Higher Eukaryotes (Shapiro and Sternberg 2005) Species Genome Size % Repetitive DNA % Coding Sequences
Table II.1 DNA content in higher eukaryotes (Shapiro and Sternberg 2005) Species Genome size1 % repetitive % coding Reference DNA sequences Animals Caenorhabditis elegans 100 MB 16.5 14 (Stein, Bao et al. 2003) Caenorhabditis 104 MB 22.4 13 (Stein, Bao et al. 2003) briggsae Drosophila 175 MB 33.7 (female) <10 (Celniker, Wheeler et al. 2002; melanogaster Bennett, Leitch et al. 2003) ~57 (male)2 Ciona intestinalis 157MB 35 9.5 (Dehal, Satou et al. 2002) Fugu rubripes 365MB 15 9.5 (Aparicio, Chapman et al. 2002) Canis domesticus 2.4GB 31 1.45 (Kirkness, Bafna et al. 2003) Mus musculus 2.5GB 40 1.4 (Waterston, Lindblad-Toh et al. 2002) Homo sapiens 2.9 GB >50 1.2 (Lander, Linton et al. 2001) Plants Arabidopsis thaliana 125-157 MB 13-14 21 (Initiative 2000; Bennett, Leitch et al. 2003) Oryza sativa (indica) 466 MB 42 11.8 (Yu, Hu et al. 2002) Oryza sativa (Japonica) 420 MB 45 11.9 (Goff, Ricke et al. 2002) Zea mays 2.5 GB 77 1 (Meyers, Tingey et al. 2001) REFERENCES Aparicio, S., J. Chapman, et al. (2002). "Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes." Science 297(5585): 1301-1310. http://www.ncbi.nlm.nih.gov/pubmed/12142439. Bennett, M. D., I. J. Leitch, et al. (2003). "Comparisons with Caenorhabditis (approximately 100 Mb) and Drosophila (approximately 175 Mb) using flow cytometry show genome size in Arabidopsis to be approximately 157 Mb and thus approximately 25% larger than the Arabidopsis genome initiative estimate of approximately 125 Mb." Ann Bot (Lond) 91(5): 547-557. -

Epigenetic Characteristics of Human Subtelomeres Vary in Cells Utilizing the Alternative Lengthening of Telomeres (ALT) Pathway
life Article Epigenetic Characteristics of Human Subtelomeres Vary in Cells Utilizing the Alternative Lengthening of Telomeres (ALT) Pathway Shir Toubiana 1,† , Aya Tzur-Gilat 1,† and Sara Selig 1,2,* 1 Department of Genetics and Developmental Biology, Rappaport Faculty of Medicine and Research Institute, Technion, Haifa 31096, Israel; [email protected] (S.T.); [email protected] (A.T.-G.) 2 Laboratory of Molecular Medicine, Rambam Health Care Campus, Haifa 31096, Israel * Correspondence: [email protected] † Both authors contributed equally. Abstract: Most human cancers circumvent senescence by activating a telomere length maintenance mechanism, most commonly involving telomerase activation. A minority of cancers utilize the recombination-based alternative lengthening of telomeres (ALT) pathway. The exact requirements for unleashing normally repressed recombination at telomeres are yet unclear. Epigenetic modifications at telomeric regions were suggested to be pivotal for activating ALT; however, conflicting data exist regarding their exact nature and necessity. To uncover common ALT-positive epigenetic characteristics, we performed a comprehensive analysis of subtelomeric DNA methylation, histone modifications, and TERRA expression in several ALT-positive and ALT-negative cell lines. We found that subtelomeric DNA methylation does not differentiate between the ALT-positive and ALT- negative groups, and most of the analyzed subtelomeres within each group do not share common Citation: Toubiana, S.; Tzur-Gilat, A.; DNA methylation patterns. Additionally, similar TERRA levels were measured in the ALT-positive Selig, S. Epigenetic Characteristics of and ALT-negative groups, and TERRA levels varied significantly among the members of the ALT- Human Subtelomeres Vary in Cells positive group. Subtelomeric H3K4 and H3K9 trimethylation also differed significantly between Utilizing the Alternative Lengthening samples in the ALT-positive group. -

Subtelomere Organization in the Genome of the Microsporidian Encephalitozoon Cuniculi: Patterns of Repeated Sequences and Physic
Dia et al. BMC Genomics (2016) 17:34 DOI 10.1186/s12864-015-1920-7 RESEARCH ARTICLE Open Access Subtelomere organization in the genome of the microsporidian Encephalitozoon cuniculi: patterns of repeated sequences and physicochemical signatures Ndongo Dia1*, Laurence Lavie2, Ngor Faye3, Guy Méténier2, Edouard Yeramian4, Christophe Duroure5, Bhen S. Toguebaye3, Roger Frutos6, Mbayame N. Niang1, Christian P. Vivarès2, Choukri Ben Mamoun7 and Emmanuel Cornillot8,9* Abstract Background: The microsporidian Encephalitozoon cuniculi is an obligate intracellular eukaryotic pathogen with a small nuclear genome (2.9 Mbp) consisting of 11 chromosomes. Although each chromosome end is known to contain a single rDNA unit, the incomplete assembly of subtelomeric regions following sequencing of the genome identified only 3 of the 22 expected rDNA units. While chromosome end assembly remains a difficult process in most eukaryotic genomes, it is of significant importance for pathogens because these regions encode factors important for virulence and host evasion. Results: Here we report the first complete assembly of E. cuniculi chromosome ends, and describe a novel mosaic structure of segmental duplications (EXT repeats) in these regions. EXT repeats range in size between 3.5 and 23.8 kbp and contain four multigene families encoding membrane associated proteins. Twenty-one recombination sites were identified in the sub-terminal region of E. cuniculi chromosomes. Our analysis suggests that these sites contribute to the diversity of chromosome ends organization through Double Strand Break repair mechanisms. The region containing EXT repeats at chromosome extremities can be differentiated based on gene composition, GC content, recombination sites density and chromosome landscape. Conclusion: Together this study provides the complete structure of the chromosome ends of E. -

Evidence-Based Guideline: Evaluation, Diagnosis, and Management Of
Evidence-based Guideline: Evaluation, Diagnosis, and Management of Facioscapulohumeral Muscular Dystrophy Report of the Guideline Development, Dissemination, and Implementation Subcommittee of the American Academy of Neurology and the Practice Issues Review Panel of the American Association of Neuromuscular & Electrodiagnostic Medicine Rabi Tawil, MD, FAAN1; John T. Kissel, MD, FAAN2; Chad Heatwole, MD, MS-CI3; Shree Pandya, PT, DPT, MS4; Gary Gronseth, MD, FAAN5; Michael Benatar, MBChB, DPhil, FAAN6 (1) MDA Neuromuscular Disease Clinic, School of Medicine and Dentistry, University of Rochester Medical Center, Rochester, NY (2) Department of Neurology, Wexner Medical Center, Ohio State University, Columbus, OH (3) Department of Neurology, School of Medicine and Dentistry, University of Rochester Medical Center, Rochester, NY (4) Department of Neurology, School of Medicine and Dentistry, University of Rochester Medical Center, Rochester, NY (5) Department of Neurology, University of Kansas School of Medicine, Kansas City, KS (6) Department of Neurology, Miller School of Medicine, University of Miami, Miami, FL Correspondence to: American Academy of Neurology [email protected] 1 Approved by the Guideline Development, Dissemination, and Implementation Subcommittee on July 23, 2014; by the AAN Practice Committee on October 20, 2014; by the AANEM Board of Directors on [date]; and by the AANI Board of Directors on [date]. This guideline was endorsed by the FSH Society on December 18, 2014. 2 AUTHOR CONTRIBUTIONS Rabi Tawil: study concept and design, acquisition of data, analysis or interpretation of data, drafting/revising the manuscript, critical revision of the manuscript for important intellectual content, study supervision. John Kissel: acquisition of data, analysis or interpretation of data, critical revision of the manuscript for important intellectual content. -

The Repetitive Landscape of the 5100 Mbp Barley Genome Thomas Wicker1*, Alan H
Wicker et al. Mobile DNA (2017) 8:22 DOI 10.1186/s13100-017-0102-3 RESEARCH Open Access The repetitive landscape of the 5100 Mbp barley genome Thomas Wicker1*, Alan H. Schulman2,3, Jaakko Tanskanen2,3, Manuel Spannagl4, Sven Twardziok4, Martin Mascher5,6, Nathan M. Springer7, Qing Li7,8, Robbie Waugh9,10, Chengdao Li11,12, Guoping Zhang13, Nils Stein5, Klaus F. X. Mayer4,14 and Heidrun Gundlach4 Abstract Background: While transposable elements (TEs) comprise the bulk of plant genomic DNA, how they contribute to genome structure and organization is still poorly understood. Especially in large genomes where TEs make the majority of genomic DNA, it is still unclear whether TEs target specific chromosomal regions or whether they simply accumulate where they are best tolerated. Results: Here, we present an analysis of the repetitive fraction of the 5100 Mb barley genome, the largest angiosperm genome to have a near-complete sequence assembly. Genes make only about 2% of the genome, while over 80% is derived from TEs. The TE fraction is composed of at least 350 different families. However, 50% of the genome is comprised of only 15 high-copy TE families, while all other TE families are present in moderate or low copy numbers. We found that the barley genome is highly compartmentalized with different types of TEs occupying different chromosomal “niches”, such as distal, interstitial, or proximal regions of chromosome arms. Furthermore, gene space represents its own distinct genomic compartment that is enriched in small non-autonomous DNA transposons, suggesting that these TEs specifically target promoters and downstream regions. -

Human Subtelomeric Duplicon Structure and Organization Comment Anthony Ambrosini*†, Sheila Paul*, Sufen Hu* and Harold Riethman*
Open Access Research2007AmbrosinietVolume al. 8, Issue 7, Article R151 Human subtelomeric duplicon structure and organization comment Anthony Ambrosini*†, Sheila Paul*, Sufen Hu* and Harold Riethman* Addresses: *The Wistar Institute, Spruce St, Philadelphia, PA 19104, USA. †Department of Molecular Biology, Princeton University, Princeton, NJ 08544, USA. Correspondence: Harold Riethman. Email: [email protected] reviews Published: 30 July 2007 Received: 29 March 2007 Revised: 25 June 2007 Genome Biology 2007, 8:R151 (doi:10.1186/gb-2007-8-7-r151) Accepted: 30 July 2007 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2007/8/7/R151 © 2007 Ambrosini et al.; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Subtelomere<p>Thetelomere sequence alleles; structure a divergenceclass of duplicon within blocks subtelomeric was identified duplicon that families are subtelomere-specific.</p> varies considerably, as does the organization of duplicon blocks at sub- reports Abstract Background: Human subtelomeric segmental duplications ('subtelomeric repeats') comprise deposited research about 25% of the most distal 500 kb and 80% of the most distal 100 kb in human DNA. A systematic analysis of the duplication substructure of human subtelomeric regions was done in order to develop a detailed understanding of subtelomeric sequence organization and a nucleotide sequence-level characterization of subtelomeric duplicon families. Results: The extent of nucleotide sequence divergence within subtelomeric duplicon families varies considerably, as does the organization of duplicon blocks at subtelomere alleles. -

Analysis of the VSG Gene Silent Archive in Trypanosoma Brucei
Downloaded from genome.cshlp.org on September 30, 2021 - Published by Cold Spring Harbor Laboratory Press Letter Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure Lucio Marcello and J. David Barry1 Wellcome Centre for Molecular Parasitology, University of Glasgow, Glasgow Biomedical Research Centre, Glasgow G12 8TA, United Kingdom Trypanosoma brucei evades host acquired immunity through differential activation of its large archive of silent variant surface glycoprotein (VSG) genes, most of which are pseudogenes in subtelomeric arrays. We have analyzed 940 VSGs, representing one half to two thirds of the arrays. Sequence types A and B of the VSG N-terminal domains were confirmed, while type C was found to be a constituent of type A. Two new C-terminal domain types were found. Nearly all combinations of domain types occurred, with some bias to particular combinations. One-third of encoded N-terminal domains, but only 13% of C-terminal domains, are intact, indicating a particular need for silent VSGs to gain a functional C-terminal domain to be expressed. About 60% of VSGs are unique, the rest occurring in subfamilies of two to four close homologs (>50%–52% peptide identity). We found a subset of VSG-related genes, differing from VSGs in genomic environment and expression patterns, and predict they have distinct function. Almost all (92%) full-length array VSGs have the partially conserved flanks associated with the duplication mechanism that activates silent genes, and these sequences have also contributed to archive evolution, mediating most of the conversions of segments, containing Ն1 VSG, within and between arrays. -

Factors That Affect Targeting of Ty5 Retrotransposon in Saccharomyces Cerevisiae Robert Dick Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 Factors that affect targeting of Ty5 retrotransposon in Saccharomyces cerevisiae Robert Dick Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Genetics and Genomics Commons Recommended Citation Dick, Robert, "Factors that affect targeting of Ty5 retrotransposon in Saccharomyces cerevisiae" (2007). Retrospective Theses and Dissertations. 15047. https://lib.dr.iastate.edu/rtd/15047 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Factors that affect targeting of Ty5 retrotransposon in Saccharomyces cerevisiae by Robert Dick A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Major: Interdepartmental Genetics Program of Study Committee: Daniel F. Voytas, Major Professor Linda Abrosio Diane Bassham Iowa State University Ames, Iowa 2007 Copyright Robert Dick, 2007. All rights reserved. UMI Number: 1446046 Copyright 2007 by Dick, Robert All rights reserved. ____________________________________________________________ UMI Microform 1446046 Copyright 2007 by ProQuest Information and Learning Company. -

The Mechanism of Integration Preference to Heterochromatin of Yeast Retrotransposon Ty5 Weiwu Xie Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2003 The mechanism of integration preference to heterochromatin of yeast retrotransposon Ty5 Weiwu Xie Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Genetics Commons, and the Molecular Biology Commons Recommended Citation Xie, Weiwu, "The mechanism of integration preference to heterochromatin of yeast retrotransposon Ty5 " (2003). Retrospective Theses and Dissertations. 1475. https://lib.dr.iastate.edu/rtd/1475 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. The mechanism of integration preference to heterochromatin of yeast retrotransposon Ty5 by Weiwu Xie A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Genetics (Computational Molecular Biology) Program of Study Committee: Daniel F. Voytas, Major Professor John May field Alan Myers David Oliver Jo Anne Powell-Coffman Iowa State University Ames, Iowa 2003 UMI Number: 3105118 UMI UMI Microform 3105118 Copyright 2003 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. Box 1346 Ann Arbor, Ml 48106-1346 ii Graduate College Iowa State University This is to certify that the doctoral dissertation of Weiwu Xie Has met the dissertation requirements of Iowa State University Signature was redacted for privacy. -

Contrasting Evolutionary Genome Dynamics Between Domesticated and Wild Yeasts
ARTICLES OPEN Contrasting evolutionary genome dynamics between domesticated and wild yeasts Jia-Xing Yue1 , Jing Li1, Louise Aigrain2, Johan Hallin1 , Karl Persson3 , Karen Oliver2, Anders Bergström2, Paul Coupland2,5, Jonas Warringer3 , Marco Cosentino Lagomarsino4, Gilles Fischer4, Richard Durbin2 & Gianni Liti1 Structural rearrangements have long been recognized as an important source of genetic variation, with implications in phenotypic diversity and disease, yet their detailed evolutionary dynamics remain elusive. Here we use long-read sequencing to generate end- to-end genome assemblies for 12 strains representing major subpopulations of the partially domesticated yeast Saccharomyces cerevisiae and its wild relative Saccharomyces paradoxus. These population-level high-quality genomes with comprehensive annotation enable precise definition of chromosomal boundaries between cores and subtelomeres and a high-resolution view of evolutionary genome dynamics. In chromosomal cores, S. paradoxus shows faster accumulation of balanced rearrangements (inversions, reciprocal translocations and transpositions), whereas S. cerevisiae accumulates unbalanced rearrangements (novel insertions, deletions and duplications) more rapidly. In subtelomeres, both species show extensive interchromosomal reshuffling, with a higher tempo in S. cerevisiae. Such striking contrasts between wild and domesticated yeasts are likely to reflect the influence of human activities on structural genome evolution. Understanding how genetic variation translates into phenotypic biology and genetics. It was the first eukaryote to have its genome diversity is a central theme in biology. With the rapid advancement of sequence, population genomics and genotype–phenotype map exten- sequencing technology, genetic variation in large natural populations sively explored1,20,21. Here we applied PacBio sequencing to 12 repre- has been explored extensively for humans and several model organ- sentative strains of S.