Wildlife in an Anthropogenically-Driven World
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

New Zealand Comprehensive II Trip Report 31St October to 16Th November 2016 (17 Days)
New Zealand Comprehensive II Trip Report 31st October to 16th November 2016 (17 days) The Critically Endangered South Island Takahe by Erik Forsyth Trip report compiled by Tour Leader: Erik Forsyth RBL New Zealand – Comprehensive II Trip Report 2016 2 Tour Summary New Zealand is a must for the serious seabird enthusiast. Not only will you see a variety of albatross, petrels and shearwaters, there are multiple- chances of getting out on the high seas and finding something unusual. Seabirds dominate this tour and views of most birds are alongside the boat. There are also several land birds which are unique to these islands: kiwis - terrestrial nocturnal inhabitants, the huge swamp hen-like Takahe - prehistoric in its looks and movements, and wattlebirds, the saddlebacks and Kokako - poor flyers with short wings Salvin’s Albatross by Erik Forsyth which bound along the branches and on the ground. On this tour we had so many highlights, including close encounters with North Island, South Island and Little Spotted Kiwi, Wandering, Northern and Southern Royal, Black-browed, Shy, Salvin’s and Chatham Albatrosses, Mottled and Black Petrels, Buller’s and Hutton’s Shearwater and South Island Takahe, North Island Kokako, the tiny Rifleman and the very cute New Zealand (South Island wren) Rockwren. With a few members of the group already at the hotel (the afternoon before the tour started), we jumped into our van and drove to the nearby Puketutu Island. Here we had a good introduction to New Zealand birding. Arriving at a bay, the canals were teeming with Black Swans, Australasian Shovelers, Mallard and several White-faced Herons. -

Developing Methods for the Field Survey and Monitoring of Breeding Short-Eared Owls (Asio Flammeus) in the UK: Final Report from Pilot Fieldwork in 2006 and 2007
BTO Research Report No. 496 Developing methods for the field survey and monitoring of breeding Short-eared owls (Asio flammeus) in the UK: Final report from pilot fieldwork in 2006 and 2007 A report to Scottish Natural Heritage Ref: 14652 Authors John Calladine, Graeme Garner and Chris Wernham February 2008 BTO Scotland School of Biological and Environmental Sciences, University of Stirling, Stirling, FK9 4LA Registered Charity No. SC039193 ii CONTENTS LIST OF TABLES................................................................................................................... iii LIST OF FIGURES ...................................................................................................................v LIST OF FIGURES ...................................................................................................................v LIST OF APPENDICES...........................................................................................................vi SUMMARY.............................................................................................................................vii EXECUTIVE SUMMARY ................................................................................................... viii CRYNODEB............................................................................................................................xii ACKNOWLEDGEMENTS....................................................................................................xvi 1. BACKGROUND AND AIMS...........................................................................................2 -

Attraction of Nocturnally Migrating Birds to Artificial Light the Influence
Biological Conservation 233 (2019) 220–227 Contents lists available at ScienceDirect Biological Conservation journal homepage: www.elsevier.com/locate/biocon Attraction of nocturnally migrating birds to artificial light: The influence of colour, intensity and blinking mode under different cloud cover conditions T ⁎ Maren Rebkea, , Volker Dierschkeb, Christiane N. Weinera, Ralf Aumüllera, Katrin Hilla, Reinhold Hilla a Avitec Research GbR, Sachsenring 11, 27711 Osterholz-Scharmbeck, Germany b Gavia EcoResearch, Tönnhäuser Dorfstr. 20, 21423 Winsen (Luhe), Germany ARTICLE INFO ABSTRACT Keywords: A growing number of offshore wind farms have led to a tremendous increase in artificial lighting in the marine Continuous and intermittent light environment. This study disentangles the connection of light characteristics, which potentially influence the Light attraction reaction of nocturnally migrating passerines to artificial illumination under different cloud cover conditions. In a Light characteristics spotlight experiment on a North Sea island, birds were exposed to combinations of light colour (red, yellow, Migrating passerines green, blue, white), intensity (half, full) and blinking mode (intermittent, continuous) while measuring their Nocturnal bird migration number close to the light source with thermal imaging cameras. Obstruction light We found that no light variant was constantly avoided by nocturnally migrating passerines crossing the sea. The number of birds did neither differ between observation periods with blinking light of different colours nor compared to darkness. While intensity did not influence the number attracted, birds were drawn more towards continuous than towards blinking illumination, when stars were not visible. Red continuous light was the only exception that did not differ from the blinking counterpart. Continuous green, blue and white light attracted significantly more birds than continuous red light in overcast situations. -

Birds: Structure, Function and Adaptation
Birds: Structure, function and adaptation Birds: Structure, function and adaptation This interactive diagram explores the sequential and interlinking science concepts that underpin knowledge and understanding about birds’ physical features, their functions and how they help birds with flight, feeding and life in particular habitats. The concepts listed just above the overarching concepts reflect learning at New Zealand Curriculum level 1 and show how they may build in sequence to level 4. The overarching science concepts are fully developed concepts and might not be achieved until level 7 or 8. Some of the text is courtesy of the New Zealand Ministry of Education’s Building Science Concepts Book 3 Birds: Structure, Function, and Adaptation. The links to Hub resources provide additional background information and classroom activities that will support teachers to scaffold the development of their students’ conceptual understanding about birds. The images provide a means to initiate discussions, check student thinking and consolidate student understanding. The article Building Science Concepts: Birds provides additional science and pedagogical information. Index • Highly specialised birds have unique adaptations that restrict them to habitats that meet their needs • Birds that are common in towns have adaptations that enable them to cope with changes brought about by people • We can usually tell what sort of food a bird eats by looking at its beak and feet • All birds have feathers, two legs and a beak instead of teeth © Copyright. Science -

Adobe PDF, Job 6
Noms français des oiseaux du Monde par la Commission internationale des noms français des oiseaux (CINFO) composée de Pierre DEVILLERS, Henri OUELLET, Édouard BENITO-ESPINAL, Roseline BEUDELS, Roger CRUON, Normand DAVID, Christian ÉRARD, Michel GOSSELIN, Gilles SEUTIN Éd. MultiMondes Inc., Sainte-Foy, Québec & Éd. Chabaud, Bayonne, France, 1993, 1re éd. ISBN 2-87749035-1 & avec le concours de Stéphane POPINET pour les noms anglais, d'après Distribution and Taxonomy of Birds of the World par C. G. SIBLEY & B. L. MONROE Yale University Press, New Haven and London, 1990 ISBN 2-87749035-1 Source : http://perso.club-internet.fr/alfosse/cinfo.htm Nouvelle adresse : http://listoiseauxmonde.multimania. -

Breeding Biology of Icelandic Thrushes
Breeding biology of Icelandic thrushes Hulda Elísabet Harðardóttir Faculty of Life and Environmental Sciences University of Iceland 2019 Breeding biology of Icelandic thrushes Hulda Elísabet Harðardóttir 90 ECTS thesis submitted in partial fulfillment of a Magister Scientiarum degree in Biology MS Committee Gunnar Þór Hallgrímsson Snæbjörn Pálsson Master’s Examiner Tómas Grétar Gunnarsson Faculty of Life and Environmental Sciences School of Engineering and Natural Sciences University of Iceland Reykjavik, May 2019 Breeding biology of Icelandic thrushes Biology of thrushes 90 ECTS thesis submitted in partial fulfillment of a Magister Scientiarum degree in Biology Copyright © 2019 Hulda Elísabet Harðardóttir All rights reserved Faculty of Life and Environmental Sciences School of Engineering and Natural Sciences University of Iceland Askja, Sturlugata 7 107, Reykjavik Iceland Telephone: 525 4000 Bibliographic information: Hulda Elísabet Harðardóttir, 2019, Breeding biology of Icelandic thrushes, Master’s thesis, Faculty of Life and Environmental Sciences, University of Iceland, pp. 68. Printing: Háskólaprent Reykjavik, Iceland, May 2019 Abstract This thesis integrates in three separate chapters some aspects of the breeding biology of the Eurasian redwing (Turdus iliacus coburni) and the Eurasian blackbird (T. merula). The redwing has been breeding in Iceland for centuries while the blackbird colonized between 1990-2000 and is still a relatively scarce breeding bird outside SW-Iceland. Both species are understudied in Iceland despite many valuable research questions, especially on interactions between the species due to the recent colonisation of the blackbird. The study was conducted in two consecutive summers in 2017 and 2018, in Fossvogur cemetery located centrally in Reykjavik Iceland. The first chapter focuses on various aspects of their breeding biology including timing, nest site selection and breeding success. -

Common Urban Birds
Common Urban Birds Crested Pigeon Spotted Turtle Dove* Feral Pigeon* Noisy Miner New Holland Eastern Spinebill White-plumed Honeyeater Honeyeater JS SW SW SW SW JS JS (Crest on head) (White spots on neck) (Dark grey feathers usually with a (Black head, yellow around eyes) (Black and yellow wings, Black and (Black, white and reddish-brown (White lines on neck) shiny green neck) white striped chest) feathers) Nectarivore & Granivore Granivore Granivore Nectarivore & Insectivore Nectarivore & Insectivore Nectarivore & Insectivore q q q q Insectivore,Omnivore q q q X Ground X Trees,Shrubs,Ground X Ground X Trees,Shrubs,Ground,Air X Trees,Shrubs,Air X Shrubs,Air X Trees,Shrubs,Ground,Air Red Wattlebird Little Wattlebird Striated Pardalote Welcome Swallow House Sparrow* Silvereye Willie Wagtail JS SW JH SW JMG JT JS (Yellow-orange belly, red wattles) (No orange on belly, no wattles) (Yellow face, black & white (Flies around ovals and other (Very small) (Silver ring around eye) (Black and white, tail wags from streaked crown, white wing streaks grassed areas, forked tail) side to side) with red spot) Nectarivore & Nectarivore & Insectivore Nectarivore & Insectivore Insectivore Granivore Omnivore Insectivore q q q Insectivore,Insectivore q q q q X Trees,Shrubs,Air X Trees,Shrubs,Air X Trees,Shrubs X Air X Ground X Trees,Shrubs X Ground,Air Common Blackbird* Common Starling* Australian Magpie Magpie-lark Little Raven Laughing Nankeen Kestrel Kookaburra JS JS JG JS JS JG JS (Smaller beak and body than (Breeding male black with bright yellow (Dark -
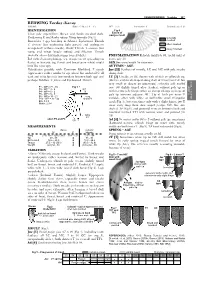
REDWING Turdus Iliacus (REDWI) Ring: 3.5 MA (3.8 – 4.2) WP = 3 (4) Incubation: F Parental Care: F, M IDENTIFICATION Fig 4 Extent of Clear Pale Supercilium
PASSERIFORMES – Turdidae 267 REDWING Turdus iliacus (REDWI) Ring: 3.5 MA (3.8 – 4.2) WP = 3 (4) Incubation: F Parental care: F, M IDENTIFICATION Fig 4 Extent of Clear pale supercilium. Breast and flanks streaked dark. postjuv moult Underwing C and flanks rufous. Wing formula (Fig 1). generally Resembles 3 spp breeding in Siberia: Eyebrowed Thrush moulted T. obscurus (but underwing light greyish and underparts often moulted orange-buff without streaks), Dusky Thrush T. e u n om u s (but rarely moulted rump and wings largely rufous) and Siberian Thrush Geokichla sibirica (but underwings barred white). PNEUMATISATION Reliable until late 09, useful until at Ind with aberrant plumage (eg, orange breast spreading on least early 10. flanks) or leucistic (eg, throat and breast pure white) might SEX See wing length for extremes. look like rarer spp. AUTUMN – AGE Hybridisation possible with Common Blackbird T. m e r u la Juv [3J] Feathers of mantle, LC and MC with pale streaks (appearance rather similar to ssp coburni but undertail C all along shaft. dark and wing formula intermediate between both spp) and 1Y [3] Usually, juv GC shorter with whitish or yellowish tip, perhaps Fieldfare T. pilaris and Eyebrowed Thrush. (distinct and streak shaped along shaft of innermost GC but very small or absent on outermost) contrasting with moulted P1 - WP = 71 - 85 Fig 1 inner GC slightly tinged olive (darker), without pale tip or TURVIS P2 - WP = 3 - 8 ≤ 8 - – P4 - WP = 0 - 2 16 with narrow pale fringe: often an abrupt change in shape of P5 - WP = 5 - 8 pale tip between adjacent GC. -

A Critical Evaluation of Neophobia in Corvids: Causes, Consequences and Conservation Implications Alison Linda Greggor King’S College
A critical evaluation of neophobia in corvids: causes, consequences and conservation implications Alison Linda Greggor King’s College October 2016 This dissertation is submitted to the University of Cambridge for the degree of Doctor of Philosophy i Preface This dissertation is the result of my own work and includes nothing which is the outcome of work done in collaboration except as detailed on the Declaration page and specified in the text. No part of this thesis has been submitted, or is being concurrently submitted, to any other university in application for a higher degree. The text does not exceed 60,000 words. ii Summary Neophobia, or the fear of novelty, is thought to restrict animals’ ecological niches and hinder their propensity for innovation; two processes that should limit behavioural adjustment to human-induced changes in the environment. However, birds within the corvid family (Corvidae) defy this trend by being highly neophobic, yet highly successful alongside humans across diverse habitats. This thesis examines the causes and ecological consequences of neophobia to unravel corvids’ puzzling neophobic tendencies. Throughout the thesis I find evidence that corvids are very neophobic, but that individuals differ in their level of novelty avoidance. Neophobia is not a fixed trait across time and towards all types of novelty. Neophobia levels differ depending on the type of novel stimuli being presented, and individuals can be inconsistent when environments change seasonally (Chapter Three). Although individual differences in neophobia are expected to be associated with fitness outcomes, I found no direct connections between neophobia, reproductive success or offspring stress hormone expression (Chapter Four). -

Volume 2, Chapter 16-2: Birds and Bryophytic Food Sources
Glime, J. M. 2017. Birds and Bryophytic Food Sources. Chapt. 16-2. In: Glime, J. M. Bryophyte Ecology. Volume 2. Bryological 16-2-1 Interaction. eBook sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 19 July 2020 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology2/>. CHAPTER 16-2 BIRDS AND BRYOPHYTIC FOOD SOURCES TABLE OF CONTENTS Capsules ........................................................................................................................................................... 16-2-2 Ptarmigans................................................................................................................................................. 16-2-5 Grouse ....................................................................................................................................................... 16-2-7 Titmice ...................................................................................................................................................... 16-2-7 Kōkako ........................................................................................ 16-2-8 Fruit Mimicry by Capsules? ...................................................................................................................... 16-2-9 Bird Color Vision ............................................................................................................................. 16-2-10 Leafy Plants ................................................................................................................................................... -

Prey Preferences and Recent Changes in Diet of a Breeding Population of the Northern Goshawk Accipiter Gentilis in Southwestern Europe
Bird Study ISSN: 0006-3657 (Print) 1944-6705 (Online) Journal homepage: http://www.tandfonline.com/loi/tbis20 Prey preferences and recent changes in diet of a breeding population of the Northern Goshawk Accipiter gentilis in Southwestern Europe Salvador Rebollo, Gonzalo García-Salgado, Lorenzo Pérez-Camacho, Sara Martínez-Hesterkamp, Alberto Navarro & José-Manuel Fernández-Pereira To cite this article: Salvador Rebollo, Gonzalo García-Salgado, Lorenzo Pérez-Camacho, Sara Martínez-Hesterkamp, Alberto Navarro & José-Manuel Fernández-Pereira (2017): Prey preferences and recent changes in diet of a breeding population of the Northern Goshawk Accipiter gentilis in Southwestern Europe, Bird Study, DOI: 10.1080/00063657.2017.1395807 To link to this article: https://doi.org/10.1080/00063657.2017.1395807 View supplementary material Published online: 20 Nov 2017. Submit your article to this journal Article views: 19 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=tbis20 Download by: [178.57.158.158] Date: 23 November 2017, At: 04:33 BIRD STUDY, 2017 https://doi.org/10.1080/00063657.2017.1395807 Prey preferences and recent changes in diet of a breeding population of the Northern Goshawk Accipiter gentilis in Southwestern Europe Salvador Rebollo a, Gonzalo García-Salgadoa, Lorenzo Pérez-Camachoa, Sara Martínez-Hesterkampa, Alberto Navarrob and José-Manuel Fernández-Pereirac aEcology and Forest Restoration Group, Department of Life Sciences, University of Alcalá, Madrid, Spain; bAsturias, Spain; cPontevedra, Spain ABSTRACT ARTICLE HISTORY Capsule: Northern Goshawk Accipiter gentilis diet has changed significantly since the 1980s, Received 2 December 2016 probably due to changes in populations of preferred prey species. -

Birding Checklist for a Tour with Wrybill Birding Tours, NZ This Checklist Covers a Single Day Tour with Brent Stephenson in Hawkes Bay
Birding checklist for a tour with Wrybill Birding Tours, NZ This checklist covers a single day tour with Brent Stephenson in Hawkes Bay. Locations visited were Westshore Lagoons and Ahuriri Estuary,Lake Tutira, and Boundary Stream Mainland Island. 18-Feb-10 Comments GREBES - PODICIPEDIDAE E New Zealand grebe (dabchick) Poliocephalus rufopectus 1 BOOBIES AND GANNETS- SULIDAE Australasian gannet Morus serrator 1 CORMORANTS - PHALACROCORACIDAE Great cormorant (black shag) Phalacrocorax carbo 1 Little black cormorant Phalacrocorax sulcirostris 1 Little pied cormorant Phalacrocorax melanoleucos 1 HERONS, EGRETS, BITTERNS - ARDEIDAE White-faced heron Egretta novaehollandiae 1 DUCKS, GEESE, SWANS - ANATIDAE Mute swan Cygnus olor 1 Black swan Cygnus atratus 1 Feral (greylag) goose Anser anser 1 Canada goose Branta canadensis 1 E Paradise shelduck Tadorna variegata 1 Mallard Anas platyrhynchos 1 Pacific black (grey) duck Anas superciliosa 1 Grey teal Anas gracilis 1 Australasian shoveler Anas rhynchotis 1 E New Zealand scaup Aythya novaeseelandiae 1 HAWKS, EAGLES, KITES - ACCIPTRIDAE Swamp (Australasian) harrier Circus approximans 1 E New Zealand falcon Falco novaeseelandiae 1 TURKEYS - MELEADRIDAE Wild turkey Meleagris gallopavo 1 NEW WORLD QUAIL - ODONTOPHORIDAE Indian peafowl (peacock) Pavo cristatus 1 California quail Callipepla californica 1 RAILS, GALLINULES, COOTS - RALLIDAE Pukeko (purple swamphen) Porphyrio melanotus 1 OYSTERCATCHERS - HAEMATOPODIDAE E South Island oystercatcher Haematopus finschi 1 AVOCETS & STILTS - RECURVIROSTRIDAE