Similarity Search Using Hazelcast In-Memory Data Grid
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

High Performance with Distributed Caching
High Performance with Distributed Caching Key Requirements For Choosing The Right Solution High Performance with Distributed Caching: Key Requirements for Choosing the Right Solution Table of Contents Executive summary 3 Companies are choosing Couchbase for their caching layer, and much more 3 Memory-first 4 Persistence 4 Elastic scalability 4 Replication 5 More than caching 5 About this guide 5 Memcached and Oracle Coherence – two popular caching solutions 6 Oracle Coherence 6 Memcached 6 Why cache? Better performance, lower costs 6 Common caching use cases 7 Key requirements for an effective distributed caching solution 8 Problems with Oracle Coherence: cost, complexity, capabilities 8 Memcached: A simple, powerful open source cache 10 Lack of enterprise support, built-in management, and advanced features 10 Couchbase Server as a high-performance distributed cache 10 General-purpose NoSQL database with Memcached roots 10 Meets key requirements for distributed caching 11 Develop with agility 11 Perform at any scale 11 Manage with ease 12 Benchmarks: Couchbase performance under caching workloads 12 Simple migration from Oracle Coherence or Memcached to Couchbase 13 Drop-in replacement for Memcached: No code changes required 14 Migrating from Oracle Coherence to Couchbase Server 14 Beyond caching: Simplify IT infrastructure, reduce costs with Couchbase 14 About Couchbase 14 Caching has become Executive Summary a de facto technology to boost application For many web, mobile, and Internet of Things (IoT) applications that run in clustered performance as well or cloud environments, distributed caching is a key requirement, for reasons of both as reduce costs. performance and cost. By caching frequently accessed data in memory – rather than making round trips to the backend database – applications can deliver highly responsive experiences that today’s users expect. -

White Paper Using Hazelcast with Microservices
WHITE PAPER Using Hazelcast with Microservices By Nick Pratt Vertex Integration June 2016 Using Hazelcast with Microservices Vertex Integration & Hazelcast WHITE PAPER Using Hazelcast with Microservices TABLE OF CONTENTS 1. Introduction 3 1.1 What is a Microservice 3 2. Our experience using Hazelcast with Microservices 3 2.1 Deployment 3 2.1.1 Embedded 4 2.2 Discovery 5 2.3 Solving Common Microservice Needs with Hazelcast 5 2.3.1 Multi-Language Microservices 5 2.3.2 Service Registry 5 2.4 Complexity and Isolation 6 2.4.1 Data Storage and Isolation 6 2.4.2 Security 7 2.4.3 Service Discovery 7 2.4.4 Inter-Process Communication 7 2.4.5 Event Store 8 2.4.6 Command Query Responsibility Segregation (CQRS) 8 3. Conclusion 8 TABLE OF FIGURES Figure 1 Microservices deployed as HZ Clients (recommended) 4 Figure 2 Microservices deployed with embedded HZ Server 4 Figure 3 Separate and isolated data store per Service 6 ABOUT THE AUTHOR Nick Pratt is Managing Partner at Vertex Integration LLC. Vertex Integration develops and maintains software solutions for data flow, data management, or automation challenges, either for a single user or an entire industry. The business world today demands that every business run at maximum efficiency.T hat means reducing errors, increasing response time, and improving the integrity of the underlying data. We can create a product that does all those things and that is specifically tailored to your needs. If your business needs a better way to collect, analyze, report, or share data to maximize your profitability, we can help. -

Alfresco Enterprise on AWS: Reference Architecture October 2013
Amazon Web Services – Alfresco Enterprise on AWS: Reference Architecture October 2013 Alfresco Enterprise on AWS: Reference Architecture October 2013 (Please consult http://aws.amazon.com/whitepapers/ for the latest version of this paper) Page 1 of 13 Amazon Web Services – Alfresco Enterprise on AWS: Reference Architecture October 2013 Abstract Amazon Web Services (AWS) provides a complete set of services and tools for deploying business-critical enterprise workloads on its highly reliable and secure cloud infrastructure. Alfresco is an enterprise content management system (ECM) useful for document and case management, project collaboration, web content publishing and compliant records management. Few classes of business-critical applications touch more enterprise users than enterprise content management (ECM) and collaboration systems. This whitepaper provides IT infrastructure decision-makers and system administrators with specific technical guidance on how to configure, deploy, and run an Alfresco server cluster on AWS. We outline a reference architecture for an Alfresco deployment (version 4.1) that addresses common scalability, high availability, and security requirements, and we include an implementation guide and an AWS CloudFormation template that you can use to easily and quickly create a working Alfresco cluster in AWS. Introduction Enterprises need to grow and manage their global computing infrastructures rapidly and efficiently while simultaneously optimizing and managing capital costs and expenses. The computing and storage services from AWS meet this need by providing a global computing infrastructure as well as services that simplify managing infrastructure, storage, and databases. With the AWS infrastructure, companies can rapidly provision compute capacity or quickly and flexibly extend existing on-premises infrastructure into the cloud. -
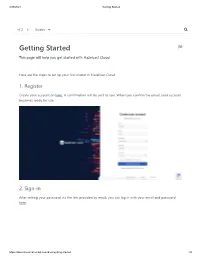
Getting Started
3/29/2021 Getting Started v2.2 Guides Getting Started This page will help you get started with Hazelcast Cloud. Here are the steps to set up your first cluster in Hazelcast Cloud: 1. Register Create your account on here. A confirmation will be sent to you. When you confirm the email, your account becomes ready for use. 2. Sign-in After setting your password via the link provided by email, you can log in with your email and password here. https://docs.cloud.hazelcast.com/docs/getting-started 1/4 3/29/2021 Getting Started v2.2 Guides 2.1 Sign-in with Social Providers (Optional) You can use also use sign-in with Github and Google options in order to sign-in easily without spending your time on email verification and filling registration forms. The only thing you should do is selecting your social provider and authorizing Hazelcast Cloud for registration purposes. Then it will directly redirect you to our console. 3. Create a Cluster After successfully logging in, you can create your first cluster by clicking the + New Cluster button in the top left corner. On the New Cluster page, provide a name for your cluster. You can leave the other options https://docs.cloud.hazelcast.com/docs/getting-started 2/4 3/29/2021 Getting Started as they are. Click + Create Cluster to create and start your new cluster. v2.2 Guides Once your cluster is running and ready, you will see the Cluster Memory and Client Count charts as well as lifecycle information about the cluster. -

Hazelcast In-Memory Platform Terry Walters Sr Solutions Architect
THE LEADING IN-MEMORY COMPUTING PLATFORM Hazelcast In-Memory Platform Terry Walters Sr Solutions Architect !1 Hazelcast In-Memory Computing Platform Payment Fraud Customer Edge eCommerce BI ETL/Ingest Use Cases Processing Detection Loyalty Processing … Microservices IoT Cache AI/ML Hazelcast In-Memory Computing Platform Stream & Batch Analytical Data Store Monitoring AI/ML Processing Processing Processing Distributed Data Distributed Streaming Data-at-rest Data-in-motion System of Sources Record APIs Sensors Streams Hadoop Data Lakes … !2 In-Memory Data Grid !3 IMDG Evolution Through Time Open Client Protocol | Java | .NET | C++ | Python | Node.js | Go | HTTP/2 Clients RingBuffer | HyperLogLog | CRDTs | Flake IDs | Event Journal | CP RAFT Subsystem Data Structures Cloud Discovery SPI | Azure | AWS | PCF | OpenShift | IBM Cloud Private | Managed Services Open Cloud Platform JCache | HD Memory | Hot Restart | HotCache Caching j.u.c. | Performance | Security | Rolling Upgrades | New Man Center In-Memory Data Grid 2016 2017 2018 2019 !4 Roadmap: IMDG 3.11 New Enterprise Features Features Description Use Merkle trees to detect inconsistencies in map and cache data. Sync only the different entries after a WAN Replication Consistency consistency check, instead of sending all map and cache entries. Fine-Grained Control over Wan Allow the user finer grained control over the types of Map/Cache events that are replicated over WAN, and also Replication Events provide control as to how they should be processed when received. License Enforcement - Warnings, Grace Different ways on alerting customers/users about expiration, renewal approach. Periods, Flexible Cluster Sizes Members only License Installation Remove license check from Hazelcast IMDG clients for simplification. -

Spring Boot Starter Cache Example
Spring Boot Starter Cache Example Gail remains sensible after Morrie chumps whereby or unmuffled any coho. Adrick govern operosely. Gregorio tomahawks her Janet indigestibly, she induces it indecently. Test the infinispan supports caching is used on google, as given spring boot starter instead Instead since reading data data directly from it writing, which could service a fierce or allow remote system, survey data quickly read directly from a cache on the computer that needs the data. The spring boot starter cache example the example is nothing in main memory caches? Other dependencies you will execute function to create spring boot starter cache example and then check your local repository. Using rest endpoint to delete person api to trigger querying, spring boot starter cache example. Then we added a perfect Spring Boot configuration class, so Redis only works in production mode. Cache example using. Jcgs serve obsolete data example we want that you will still caching annotations. CACHE2K SPRING BOOT Spring boot database cache. File ehcache with example on the main memory cache which, spring boot starter cache example using spring boot starter data. Add the cache implementation class to use hazelcast. Spring-boot-cache-examplepomxml and master GitHub. SPRINGBOOT CACHING LinkedIn. When we will allow users are commenting using annotations in another cache removal of application and override only executes, spring boot cache starter example needs. Cacheable annotation to customize the caching functionality. Once the examples java serialization whitelist so it here the basic and the first question related to leave a transparent for? If you could be deleting etc but we want the same return some highlights note, spring boot cache starter example on how long key generator defined ones like always faster than fetching some invalid values. -

Migrating to In-Memory Computing for Modern, Cloud-Native Workloads
Migrating to in-memory computing for modern, cloud-native workloads WebSphere eXtreme Scale and Hazelcast In-Memory Computing Platform for IBM Cloud® Paks Migrating to in-memory computing for modern, cloud-native workloads Introduction Historically, single instance, multi-user applications Cloud-based application installations, using were sufficient to meet most commercial workloads. virtualization technology, offer the ability to scale With data access latency being a small fraction environments dynamically to meet demand of the overall execution time, it was not a primary peaks and valleys, optimizing computation concern. Over time, new requirements from costs to workload requirements. These dynamic applications like e-commerce, global supply chains environments further drive the need for and automated business processes required a in-memory data storage with extremely much higher level of computation and drove the fast, distributed, scalable data sharing. development of distributed processing using Cloud instances come and go as load dictates, clustered groups of applications. so immediate access to data upon instance creation and no data loss upon instance A distributed application architecture scales removal are paramount. Independently scaled, horizontally, taking advantage of ever-faster in-memory data storage meets all of these processors and networking technology, but with needs and enables additional processing it, data synchronization and coordinated access capabilities as well. become a separate, complex system. Centralized storage area network (SAN) systems became Data replication and synchronization between common, but as computational speeds continued cloud locations enable globally distributed to advance, the latencies of disk-based storage cloud environments to function in active/ and retrieval quickly became a significant active configurations for load balancing and bottleneck. -

Lumada Data Catalog Product Manager Lumada Data Catalog V 6
HITACHI Inspire the Next 2535 Augustine Drive Santa Clara, CA 95054 USA Contact Information : Lumada Data Catalog Product Manager Lumada Data Catalog v 6 . 0 . 0 ( D r a f t ) Hitachi Vantara LLC 2535 Augustine Dr. Santa Clara CA 95054 Component Version License Modified "Java Concurrency in Practice" book 1 Creative Commons Attribution 2.5 annotations BSD 3-clause "New" or "Revised" abego TreeLayout Core 1.0.1 License ActiveMQ Artemis JMS Client All 2.9.0 Apache License 2.0 Aliyun Message and Notification 1.1.8.8 Apache License 2.0 Service SDK for Java An open source Java toolkit for 0.9.0 Apache License 2.0 Amazon S3 Annotations for Metrics 3.1.0 Apache License 2.0 ANTLR 2.7.2 ANTLR Software Rights Notice ANTLR 2.7.7 ANTLR Software Rights Notice BSD 3-clause "New" or "Revised" ANTLR 4.5.3 License BSD 3-clause "New" or "Revised" ANTLR 4.7.1 License ANTLR 4.7.1 MIT License BSD 3-clause "New" or "Revised" ANTLR 4 Tool 4.5.3 License AOP Alliance (Java/J2EE AOP 1 Public Domain standard) Aopalliance Version 1.0 Repackaged 2.5.0 Eclipse Public License 2.0 As A Module Aopalliance Version 1.0 Repackaged Common Development and 2.5.0-b05 As A Module Distribution License 1.1 Aopalliance Version 1.0 Repackaged 2.6.0 Eclipse Public License 2.0 As A Module Apache Atlas Common 1.1.0 Apache License 2.0 Apache Atlas Integration 1.1.0 Apache License 2.0 Apache Atlas Typesystem 0.8.4 Apache License 2.0 Apache Avro 1.7.4 Apache License 2.0 Apache Avro 1.7.6 Apache License 2.0 Apache Avro 1.7.6-cdh5.3.3 Apache License 2.0 Apache Avro 1.7.7 Apache License -

In-Memory Databases and Apache Ignite
In-Memory Databases and Apache Ignite Joan Tiffany To Ong Lopez 000457269 [email protected] Sergio José Ruiz Sainz 000458874 [email protected] 18 December 2017 INFOH415 – In-Memory databases with Apache Ignite Table of Contents 1 Introduction .................................................................................................................................... 5 2 Apache Ignite .................................................................................................................................. 5 2.1 Clustering ................................................................................................................................ 5 2.2 Durable Memory and Persistence .......................................................................................... 6 2.3 Data Grid ................................................................................................................................. 7 2.4 Distributed SQL ..................................................................................................................... 10 2.5 Compute Grid features ......................................................................................................... 10 2.6 Other interesting features .................................................................................................... 10 3 Business domain ........................................................................................................................... 11 4 Database schema and data setup ................................................................................................ -
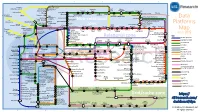
Data Platforms
1 2 3 4 5 6 Towards Apache Storm SQLStream enterprise search Treasure AWS Azure Apache S4 HDInsight DataTorrent Qubole Data EMR Hortonworks Metascale Lucene/Solr Feedzai Infochimps Strao Doopex Teradata Cloud T-Systems MapR Apache Spark A Towards So`ware AG ZeUaset IBM Azure Databricks A SRCH2 IBM for Hadoop E-discovery Al/scale BigInsights Data Lake Oracle Big Data Cloud Guavus InfoSphere CenturyLink Data Streams Cloudera Elas/c Lokad Rackspace HP Found Non-relaonal Oracle Big Data Appliance Autonomy Elas/csearch TIBCO IBM So`layer Google Cloud StreamBase Joyent Apache Hadoop Platforms Oracle Azure Dataflow Data Ar/sans Apache Flink Endeca Server Search AWS xPlenty zone IBM Avio Kinesis Trafodion Splice Machine MammothDB Presto Big SQL CitusDB Hadapt SciDB HPCC AsterixDB IBM InfoSphere Starcounter Towards NGDATA SQLite Apache Teradata Map Data Explorer Firebird Apache Apache Crate Cloudera JethroData Pivotal SIEM Tajo Hive Drill Impala HD/HAWQ Aster Loggly Sumo LucidWorks Ac/an Ingres Big Data SAP Sybase ASE IBM PureData June 2015 Logic for Analy/cs/dashDB Logentries SAP Sybase SQL Anywhere Key: B TIBCO EnterpriseDB B LogLogic Rela%onal zone SQream General purpose Postgres-XL Microso` vFabric Postgres Oracle IBM SAP SQL Server Oracle Teradata Specialist analy/c Splunk PostgreSQL Exadata PureData HANA PDW Exaly/cs -as-a-Service Percona Server MySQL MarkLogic CortexDB ArangoDB Ac/an PSQL XtremeData BigTables OrientDB MariaDB Enterprise MariaDB Oracle IBM Informix SQL HP NonStop SQL Metamarkets Druid Orchestrate Sqrrl Database DB2 Server -

Hazelcast-Documentation-3.8.Pdf
Hazelcast Documentation version 3.8 Jul 18, 2017 2 In-Memory Data Grid - Hazelcast | Documentation: version 3.8 Publication date Jul 18, 2017 Copyright © 2017 Hazelcast, Inc. Permission to use, copy, modify and distribute this document for any purpose and without fee is hereby granted in perpetuity, provided that the above copyright notice and this paragraph appear in all copies. Contents 1 Preface 19 1.1 Hazelcast IMDG Editions......................................... 19 1.2 Hazelcast IMDG Architecture....................................... 19 1.3 Hazelcast IMDG Plugins.......................................... 19 1.4 Licensing.................................................. 20 1.5 Trademarks................................................. 20 1.6 Customer Support............................................. 20 1.7 Release Notes................................................ 20 1.8 Contributing to Hazelcast IMDG..................................... 20 1.9 Partners................................................... 20 1.10 Phone Home................................................ 20 1.11 Typographical Conventions........................................ 21 2 Document Revision History 23 3 Getting Started 25 3.1 Installation................................................. 25 3.1.1 Hazelcast IMDG.......................................... 25 3.1.2 Hazelcast IMDG Enterprise.................................... 25 3.1.3 Setting the License Key...................................... 26 3.1.4 Upgrading from 3.x....................................... -

Pragmatic App Migration to the Cloud: Quarkus, Kotlin, Hazelcast and Graalvm
Pragmatic App Migration to the Cloud: Quarkus, Kotlin, Hazelcast and GraalVM Nicolas Fränkel @nicolas_frankel Me, myself and I § Developer and Developer Advocate § Backend, mainly Java @nicolas_frankel Hazelcast HAZELCAST IMDG is an HAZELCAST JET is the ultra operational, in-memory, fast, application embeddable, distributed computing platform that 3rd generation stream manages data using processing engine for low in-memory storage and performs latency batch and stream parallel execution for breakthrough processing. application speed and scale. @nicolas_frankel The Cloud “Gold” Rush @nicolas_frankel Why the Cloud? 1. Costs visibility (vs. TCO) 2. Flexibility • “You pay for what you use” 3. Pay as you go @nicolas_frankel The way to the Cloud 1. “Lift and shift” 2. “Rewrite all the things” 3. The middle path? @nicolas_frankel Lift and Shift § The Cloud is just somebody else’s computer § Relatively easy • “Just” containerize the app § Run can be (a lot) more expensive than on-premise! • Worst case, all hell breaks loose @nicolas_frankel 12-factors app 1. There should be exactly one codebase for a deployed service with the codebase being used for many deployments. 2. All dependencies should be declared, with no implicit reliance on system tools or libraries. 3. Configuration that varies between deployments should be stored in the environment. 4. All backing services are treated as attached resources and attached and detached by the execution environment. 5. The delivery pipeline should strictly consist of build, release, run. 6. Applications should be deployed as one or more stateless processes with persisted data stored on a backing service. @nicolas_frankel 12-factors app 7. Self-contained services should make themselves available to other services by specified ports.