Speciation and Phylogeography of Giant Petrels Macronectes Q
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Print 04/02 April
From the Rarities Committee’s files: Rare seabirds and a record of Herald Petrel Ian Lewington ABSTRACT Rare seabirds are often extremely hard to identify, and a significant part of the problem is that, when observed from land, circumstances are typically very difficult. In many cases, one or more of the following drawbacks applies: the weather conditions are poor, views are distant and brief, and photographic evidence is impossible. For these same reasons, records of rare seabirds are also difficult to assess, particularly so if they concern what would be a ‘first for Britain’ for the species in question.This was the case when a probable Herald Petrel Pterodroma arminjoniana was seen off Dungeness, Kent, in January 1998. In this paper, the circumstances and the assessment of that record are described, and, more generally, the level of supporting evidence which is necessary for acceptance of records of rare seabirds is discussed. 156 © British Birds 95 • April 2002 • 156-165 Rare seabirds and a record of Herald Petrel are seabirds present difficulties in many panic was beginning to set in. Had we missed ways. They are difficult to find, and most it? A few seconds later, the mystery seabird Robservers will spend hundreds of hours came into our field of view, trailing behind a ‘sifting through’ common species before Northern Gannet Morus bassanus and flying encountering a rarity. They are difficult to iden- steadily west, low over the water, about 400 m tify, not least because the circumstances in offshore. which they are seen usually mean that, com- At the time of the observation the light was pared with most other birding situations, views dull but clear, in fact excellent for observing are both distant and brief, and the observer is colour tones. -

Conservation Planning and the IUCN Red List
Vol. 6: 113–125, 2008 ENDANGERED SPECIES RESEARCH Printed December 2008 doi: 10.3354/esr00087 Endang Species Res Published online May 7, 2008 Contribution to the Theme Section ‘The IUCN Red List of Threatened Species: assessing its utility and value’ OPENPEN ACCESSCCESS REVIEW Conservation planning and the IUCN Red List M. Hoffmann1, 2,*, T. M. Brooks1, 3, 4, G. A. B. da Fonseca5, 6, C. Gascon 7, A. F. A. Hawkins7, R. E. James8, P. Langhammer9, R. A. Mittermeier7, J. D. Pilgrim10, A. S. L. Rodrigues11, J. M. C. Silva12 1Center for Applied Biodiversity Science, Conservation International, 2011 Crystal Drive Suite 500, Arlington, Virginia 22202, USA 2IUCN Species Programme, IUCN — International Union for the Conservation of Nature, Rue Mauverney, 1196 Gland, Switzerland 3World Agroforestry Center (ICRAF), University of the Philippines Los Baños, Laguna 4031, Philippines 4School of Geography and Environmental Studies, University of Tasmania, Hobart, Tasmania 7001, Australia 5Global Environment Facility, 1818 H Street NW, Washington, DC 20433, USA 6Departamento de Zoologia, Universidade Federal de Minas Gerais, Avenida Antonio Carlos 6627, Belo Horizonte MG 31270-901, Brazil 7Conservation International, 2011 Crystal Drive Suite 500, Arlington, Virginia 22202, USA 8Conservation International Melanesia Centre for Biodiversity Conservation, PO Box 106, Waigani, NCD, Papua New Guinea 9School of Life Sciences, Arizona State University, PO Box 874501, Tempe, Arizona 85287-4501, USA 10BirdLife International in Indochina, N6/2+3, Ngo 25, Lang Ha, Ba Dinh, Hanoi, Vietnam 11Department of Zoology, University of Cambridge, Cambridge CB2 3EJ, UK 12Conservation International — Brazil, Av. Gov. José Malcher 652, 2o. Andar, Ed. CAPEMI, Bairro: Nazaré, 66035-100, Belém, Pará, Brazil ABSTRACT: Systematic conservation planning aims to identify comprehensive protected area net- works that together will minimize biodiversity loss. -

Proposal for Inclusion of the Antipodean Albatross in Appendix I
CONVENTION ON UNEP/CMS/COP13/Doc. 27.1.7 MIGRATORY 25 September 2019 Original: English SPECIES 13th MEETING OF THE CONFERENCE OF THE PARTIES Gandhinagar, India, 17 - 22 February 2020 Agenda Item 27.1 PROPOSAL FOR THE INCLUSION OF THE ANTIPODEAN ALBATROSS (Diomedea antipodensis) ON APPENDIX I OF THE CONVENTION Summary: The Governments of New Zealand, Australia and Chile have submitted the attached proposal for the inclusion of the Antipodean albatross (Diomedea antipodensis) on Appendix I of CMS. The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CMS Secretariat (or the United Nations Environment Programme) concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. The responsibility for the contents of the document rests exclusively with its author. UNEP/CMS/COP13/Doc. 27.1.7 PROPOSAL FOR INCLUSION OF THE ANTIPODEAN ALBATROSS (Diomedea antipodensis) ON APPENDIX I OF THE CONVENTION A. PROPOSAL Inclusion of Diomedea antipodensis on the Convention on the Conservation of Migratory Species of Wild Animals (CMS) Appendix I. The current CMS Appendix II listing will remain in place. Diomedea antipodensis is classified as Endangered (IUCN) as it is undergoing a very rapid decline in population size. B. PROPONENT: Governments of New Zealand, Australia and Chile. C. SUPPORTING STATEMENT 1. Taxonomy 1.1 Class: Aves 1.2 Order: Procellariiformes 1.3 Family: Diomedeidae (albatrosses) 1.4 Genus, species or subspecies, including author and year: Diomedea antipodensis (Robertson & Warham 1992), including two subspecies: Diomedea antipodensis antipodensis and Diomedea antipodensis gibsoni 1.5 Scientific synonyms: Diomedea exulans antipodensis Diomedea antipodensis was formerly included in the wandering albatross complex (Diomedea exulans) (e.g. -

Pterodromarefs V1-5.Pdf
Index The general order of species follows the International Ornithological Congress’ World Bird List. A few differences occur with regard to the number and treatment of subspecies where some are treated as full species. Version Version 1.5 (5 May 2011). Cover With thanks to Kieran Fahy and Dick Coombes for the cover images. Species Page No. Atlantic Petrel [Pterodroma incerta] 5 Barau's Petrel [Pterodroma baraui] 17 Bermuda Petrel [Pterodroma cahow] 11 Black-capped Petrel [Pterodroma hasitata] 12 Black-winged Petrel [Pterodroma nigripennis] 18 Bonin Petrel [Pterodroma hypoleuca] 19 Chatham Islands Petrel [Pterodroma axillaris] 19 Collared Petrel [Pterodroma brevipes] 20 Cook's Petrel [Pterodroma cookii] 20 De Filippi's Petrel [Pterodroma defilippiana] 20 Desertas Petrel [Pterodroma deserta] 11 Fea's Petrel [Pterodroma feae] 8 Galapágos Petrel [Pterodroma phaeopygia] 17 Gould's Petrel [Pterodroma leucoptera] 19 Great-winged Petrel [Pterodroma macroptera] 3 Grey-faced Petrel [Pterodroma gouldi] 4 Hawaiian Petrel [Pterodroma sandwichensis] 17 Henderson Petrel [Pterodroma atrata] 16 Herald Petrel [Pterodroma heraldica] 14 Jamaica Petrel [Pterodroma caribbaea] 13 Juan Fernandez Petrel [Pterodroma externa] 13 Kermadec Petrel [Pterodroma neglecta] 14 Magenta Petrel [Pterodroma magentae] 6 Mottled Petrel [Pterodroma inexpectata] 18 Murphy's Petrel [Pterodroma ultima] 6 Phoenix Petrel [Pterodroma alba] 16 Providence Petrel [Pterodroma solandri] 5 Pycroft's Petrel [Pterodroma pycrofti] 21 Soft-plumaged Petrel [Pterodroma mollis] 7 Stejneger's Petrel [Pterodroma longirostris] 21 Trindade Petrel [Pterodroma arminjoniana] 15 Vanuatu Petrel [Pterodroma occulta] 13 White-headed Petrel [Pterodroma lessonii] 4 White-necked Petrel [Pterodroma cervicalis] 18 Zino's Petrel [Pterodroma madeira] 9 1 General Bailey, S.F. et al 1989. Dark Pterodroma petrels in the North Pacific: identification, status, and North American occurrence. -

Andrea Milković New Zealand and Its Tourism Potential
New Zealand and its Tourism Potential Milković, Andrea Undergraduate thesis / Završni rad 2017 Degree Grantor / Ustanova koja je dodijelila akademski / stručni stupanj: Polytechnic of Međimurje in Čakovec / Međimursko veleučilište u Čakovcu Permanent link / Trajna poveznica: https://urn.nsk.hr/urn:nbn:hr:110:471894 Rights / Prava: In copyright Download date / Datum preuzimanja: 2021-09-30 Repository / Repozitorij: Polytechnic of Međimurje in Čakovec Repository - Polytechnic of Međimurje Undergraduate and Graduate Theses Repository MEĐIMURSKO VELEUČILIŠTE U ČAKOVCU STRUČNI STUDIJ MENADŢMENT TURIZMA I SPORTA ANDREA MILKOVIĆ NEW ZEALAND AND ITS TOURISM POTENTIAL ZAVRŠNI RAD ČAKOVEC, 2016. POLYTECHNIC OF MEĐIMURJE IN ČAKOVEC PROFESSIONAL STUDY PROGRAME MANAGEMENT OF TOURISM AND SPORT ANDREA MILKOVIĆ NEW ZEALAND AND ITS TOURISM POTENTIAL FINAL PAPER Mentor: Marija Miščančuk, prof. ČAKOVEC, 2016 Zahvala: Veliku zahvalnost, u prvom redu, dugujem svojoj mentorici, prof. Mariji Miščančuk zbog savjetovanja, usmjeravanja i odvojenog vremena tijekom pisanja ovog završnog rada. Zahvaljujem se i ostalim djelatnicima na MeĎimurskom Veleučilištu u Čakovcu zbog kvalitetnog prenošenja znanja i pomoći tijekom studiranja. Veliko hvala Antoniju Kovačeviću i sestri Nikolini Milković na pomoći oko nabavljanja literature i tehničkoj podršci. Isto tako, zahvaljujem im se na ohrabrenju i moralnoj podršci za vrijeme pisanja rada, ali i tijekom cijelog studiranja. TakoĎer, hvala mojim prijateljima Goranu Haramasu, Martini Šestak, Petri Benotić, Petri Kozulić i Vinki Kugelman koji su bili uz mene i učinili ove studijske godine ljepšima. Hvala mojoj obitelji na podršci i strpljenju tokom studija. ABSTRACT Curiosity of people leads to traveling for pleasure to new places where they can visit and learn about historical buildings, natural beauty and anything that makes one country special, interesting and worth visiting. -

Potential for Rat Predation to Cause Decline of the Globally Threatened
Vol. 11: 47–59, 2010 ENDANGERED SPECIES RESEARCH Published online March 10 doi: 10.3354/esr00249 Endang Species Res OPENPEN ACCESSCCESS Potential for rat predation to cause decline of the globally threatened Henderson petrel Pterodroma atrata: evidence from the field, stable isotopes and population modelling M. de L. Brooke1,*, T. C. O’Connell2, David Wingate3, Jeremy Madeiros3, Geoff M. Hilton4, 6, Norman Ratcliffe5 1Department of Zoology, University of Cambridge, Downing Street, Cambridge CB2 3EJ, UK 2McDonald Institute for Archaeological Research, University of Cambridge, Downing Street, Cambridge CB2 3ER, UK 3Department of Conservation Services, Ministry of the Environment, PO Box FL117, Flatts, Bermuda 4Royal Society for the Protection of Birds, The Lodge, Sandy, Bedfordshire SG19 2DL, UK 5British Antarctic Survey, High Cross, Cambridge CB3 0ET, UK 6Wildfowl and Wetlands Trust, Slimbridge, Gloucestershire GL2 7BT, UK ABSTRACT: Past studies have indicated that Pacific rats Rattus exulans are significant predators of the chicks of surface-breeding seabirds, namely gadfly petrels Pterodroma spp., on Henderson Island, central South Pacific. Further fieldwork in 2003 confirmed the heavy predation of chicks of Murphy’s petrel P. ultima by rats. By extension, heavy predation is also likely each year on the endan- gered Henderson petrel P. atrata, for which Henderson Island is the only confirmed breeding site. To assess how important petrels are in the overall diet of rats, we conducted stable isotope analyses of rats from the shore, where petrels are most concentrated, and from about 1 km inland, where fewer nest. The carbon isotope results suggested that inland rats obtain about 30% of their food from marine sources, while the figure for shore rats was about 40%. -

New Zealand Subantarctic Islands Research Strategy
New Zealand Subantarctic Islands Research Strategy SOUTHLAND CONSERVANCY New Zealand Subantarctic Islands Research Strategy Carol West MAY 2005 Cover photo: Recording and conservation treatment of Butterfield Point fingerpost, Enderby Island, Auckland Islands Published by Department of Conservation PO Box 743 Invercargill, New Zealand. CONTENTS Foreword 5 1.0 Introduction 6 1.1 Setting 6 1.2 Legal status 8 1.3 Management 8 2.0 Purpose of this research strategy 11 2.1 Links to other strategies 12 2.2 Monitoring 12 2.3 Bibliographic database 13 3.0 Research evaluation and conditions 14 3.1 Research of benefit to management of the Subantarctic islands 14 3.2 Framework for evaluation of research proposals 15 3.2.1 Research criteria 15 3.2.2 Risk Assessment 15 3.2.3 Additional points to consider 16 3.2.4 Process for proposal evaluation 16 3.3 Obligations of researchers 17 4.0 Research themes 18 4.1 Theme 1 – Natural ecosystems 18 4.1.1 Key research topics 19 4.1.1.1 Ecosystem dynamics 19 4.1.1.2 Population ecology 20 4.1.1.3 Disease 20 4.1.1.4 Systematics 21 4.1.1.5 Biogeography 21 4.1.1.6 Physiology 21 4.1.1.7 Pedology 21 4.2 Theme 2 – Effects of introduced biota 22 4.2.1 Key research topics 22 4.2.1.1 Effects of introduced animals 22 4.2.1.2 Effects of introduced plants 23 4.2.1.3 Exotic biota as agents of disease transmission 23 4.2.1.4 Eradication of introduced biota 23 4.3 Theme 3 – Human impacts and social interaction 23 4.3.1 Key research topics 24 4.3.1.1 History and archaeology 24 4.3.1.2 Human interactions with wildlife 25 4.3.1.3 -

Northern Giant Petrel Macronectes Halli Breeding Population Survey, Auckland Islands
Northern giant petrel Macronectes halli breeding population survey, Auckland Islands December 2015 – February 2016 Graham C. Parker, Chris G. Muller and Kalinka Rexer‐Huber Department of Conservation, Conservation Services Programme, Contract 4655‐4 1 Northern giant petrel Macronectes halli breeding population survey, Auckland Islands December 2015– February 2016 Department of Conservation, Conservation Services Programme, Contract 4655‐4 Graham C. Parker1*, Chris G. Muller3 & Kalinka Rexer‐Huber1,2 1 Parker Conservation, 126 Maryhill Tce, Dunedin New Zealand 2 University of Otago, PO Box 56, Dunedin New Zealand 3Massey University Manawatu, Private Bag 11 222, Palmerston North 4442, New Zealand *Corresponding author: [email protected] Please cite as: Parker, G.C., Muller, C.G., Rexer‐Huber, K. 2016. Northern giant petrel Macronectes halli breeding population survey, Auckland Islands, December 2015 – February 2016. Report to the Conservation Services Programme, Department of Conservation. Parker Conservation, Dunedin 2 Executive summary Northern giant petrels Macronectes halli are a large, southern hemisphere fulmarine petrel that face conservation threats both in the terrestrial and marine environment. Introduced mammalian predators at breeding sites cause nesting failures and in some instances may also depredate adults. In the marine environment Northern giant petrels are threatened by capture in longline and trawl fisheries, oil pollution, shooting by fishers for bait stealing and the effects of climate change. The contemporary size and the population trends of Northern giant petrels on New Zealand islands are not known. Records of their numbers in the Auckland Islands are based solely on anecdotal evidence, and the most recent summary dates to the 1980s. We estimated the size of the Northern giant petrel breeding population and describe their spatial distribution in the Auckland Islands. -

Draft Programmatic Environmental Impact Statement October 27, 2005
Draft Programmatic Environmental Impact Statement Towards an Ecosystem Approach for the Western Pacific Region: From Species-based Fishery Management Plans to Place-based Fishery Ecosystem Plans October 27, 2005 Draft Programmatic Environmental Impact Statement Towards an Ecosystem Approach for the Western Pacific Region: From Species-based Fishery Management Plans to Place-based Fishery Ecosystem Plans October 27, 2005 ______________________________________________________________________________ Responsible Agency: National Marine Fisheries Service Pacific Islands Region 1601 Kapiolani Blvd., Ste. 1101 Honolulu, HI 96814 (808) 944-2200 Responsible Official: William L. Robinson Regional Administrator For Further Information Contact: William L. Robinson Kitty M. Simonds NMFS- Pacific Islands Region Western Pacific Fishery Managemnt Council 1601 Kapiolani Blvd., Suite 1110 1164 Bishop St., Ste. 1400 Honolulu, HI. 96814-4700 Honolulu, HI 96813 (808) 944-2200 ph. (808) 973-2941 fax (808) 522-8220 ph. (808) 522-8226 fax Abstract: This document analyzes the impacts on the human environment resulting from step one of the implementation of an ecosystem approach to fisheries management in the Western Pacific Region (American Samoa, Guam, Hawaii, the Commonwealth of the Northern Mariana Islands, and the U.S. Pacific Remote Island Areas1). The alternatives analyzed in this document are linked to the following five issues: the establishment of fishery ecosystem plan boundaries; the determination of appropriate management unit species; modifications -

Managementframeworksf
Papers and Proceedings ofthe Royal Society of Tasmania, Volume 141(1), 2007 29 MANAGEMENTFRAMEWORKSFOR THE NEWZEALAND SUB-ANTARCTIC ISLANDS by A. D. Roberts Roberts, A.D. 2007 (23:xi): Management frameworks for the New Zealand sub-Antarctic islands. Papers and Proceedings of the Royal Society of Tasmania 141(1): 29-32. https://doi.org/10.26749/rstpp.141.1.29 ISSN 0080-4703. Department of Conservation, PO Box 743, Invercargill, New Zealand. Email: [email protected] Thefive island groups known as the New Zealand sub-Antarctic islands - Antipodes Islands, Auckland Islands, Bounty Islands, Campbell Island and Snares Islands - are perhaps better regarded as cool temperate although they share many features and many management issues with islands south of the Antarctic Convergence. The Snares and Bounty islands have granitic substrates; the Antipodes, Auckland and Campbell islands are of volcanic origin. Marine mammals and seabirds are important components of the fauna. Terrestrial Hora and inver tebrate fauna show high levels of endemism. Several species of alien vertebrates have been successfully eradicated. The islands are managed by the New Zealand Department of Conservation as National Nature Reserves, and together are recognised as a World Heritage Site. Key Words: sub-Antarctic islands, cool temperate, Antipodes Islands, Auckland Islands, Bounty Islands, Campbell Island, Snares Islands, World Heritage Site, alien species removal. TheNew Zealand sub-Antarctic islands are located south of There is evidence of Polynesian v1s1tation in the pre New Zealand in the Pacific sector of the Southern Ocean. European period (Peat 2003). European discovery was in Although these islands are not truly sub-Antarctic but more 1791. -
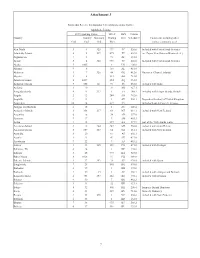
Attachment 3
Attachment 3 Points that Receive International Telecommunications Traffic: Alphabetic Listing FCC reporting Codes ITU-T FIPS Census Country Country Summary Dialing 10-3 Schedule C Comments, including other Code Code Code Plan names commonly used Abu Dhabi 3 1 325 971 TC 520.0 Included with United Arab Emirates Admiralty Islands 8 2 237 675 PP 604.0 inc. Papua New Guinea (Bismarck A.) Afghanistan 7 3 93 AF 531.0 Ajman 3 4 325 971 TC 520.0 Included with United Arab Emirates Alaska 5 1005 1 US 100.0 Albania 9 6 355 AL 481.0 Alderney 1 7 326 44 GK 412.0 Guernsey (Channel Islands) Algeria 2 8 213 AG 721.0 American Samoa 8 1009 684 AQ 951.0 Andaman Islands 7 355 141 91 IN 533.0 included with India Andorra 1 10 33 AN 427.1 Anegada Islands 4 11 337 1 VI 248.2 included with Virgin Islands, British Angola 2 12 244 AO 762.0 Anguilla 4 13 1 AV 248.1 Dependent territory of United Kingdom Antarctica 10 14 672 AY Includes Scott & Casey U.S. bases Antigua and Barbuda 4 15 1 AC 248.4 Antipodes Islands 8 356 217 64 NZ 614.1 included with New Zealand Argentina 6 16 54 AR 357.0 Armenia 9 17 7 AM 463.1 Aruba 4 18 297 AA 277.9 part of the Netherlands realm Ascension Island 2 19 264 247 SH 758.0 included with Saint Helena Auckland Islands 8 357 217 64 NZ 614.1 included with New Zealand Australia 8 20 61 AS 602.1 Austria 1 21 43 AU 433.0 Azerbaijan 9 22 7 AJ 463.2 Azores 1 23 245 351 PO 471.0 included with Portugal Bahamas, The 4 24 1 BF 236.0 Bahrain 3 25 973 BA 525.0 Baker Island 8 1026 11 FQ 980.0 Balearic Islands 1 27 293 34 SP 470.0 included with Spain -

Herald Petrel New to the West Indies
EXTRALIMITAL RECORD Herald Petrel new to the West Indies Michael Gochfeld,Joanna Burger, Jorge Saliva, and Deborah Gochfeld (Pterodroma)are bestrepresented SinA southGROUP temperate THEGADFLY and subantarc-PETRELS tic waterswith a few speciesbreeding on tropicalislands in the Central Pacific, Atlantic, and Indian Oceans.In the West Indies,the only member of this genusis the Black-cappedPetrel (P hasitata), which breeds in the Greater Antilles and perhaps still in the Lesser Antilles. A dose relative,the endangeredBermuda (P..cahow) breeds in Bermuda. On July 12, 1986, while studyingthe breeding Laughing Gulls (Larus atri- cilia), Royal Terns,and SandwichTerns (Sterna maxima and S. sandvicensis)on Cayo Lobito, in the Culebra National Wildlife Refuge, about 40 kilometers eastof Fajardo,Puerto Rico, we flushed an all-dark petrel from a scrapeamong the nestingterns. As soon as the bird wheeled around we realized that it was a petrel, and not the familiar Sooty Shearwater(Puj•nus griseus),a species Figure 1. Dark-phasePterodroma landing among Laughing Gulls on Cayo Lobito. Culebra, which would itselfbe extremelyunusual PuertoRico. Note the ashygray undersurface of theprimary.feathers, ending in longnarrow at Culebra. Compared to a shearwater, "digital" extensionsthat are the pale areas of the inner websof theprimaries. The greater the petrelappeared to havea largerhead underwingcoverts are pale with a narrowdark tip that showsas thefaint wavybar at the base of theprimaries. The Iongishtail and shortishbill are apparent. and a longerand slightlywedge-shaped tail. It did not showthe conspicuouspale wing linings of the Sooty Shearwater (Fig. 1). The bird circled rapidly, first low over the water and then wheeling high overthe rocky islet,before return- ing to its scrapeamong the gulls and terns.