Purnama A. A., Mubarak J., Daruwati I., Roslim D. I., Elvyra R., 2019 First
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

§4-71-6.5 LIST of CONDITIONALLY APPROVED ANIMALS November
§4-71-6.5 LIST OF CONDITIONALLY APPROVED ANIMALS November 28, 2006 SCIENTIFIC NAME COMMON NAME INVERTEBRATES PHYLUM Annelida CLASS Oligochaeta ORDER Plesiopora FAMILY Tubificidae Tubifex (all species in genus) worm, tubifex PHYLUM Arthropoda CLASS Crustacea ORDER Anostraca FAMILY Artemiidae Artemia (all species in genus) shrimp, brine ORDER Cladocera FAMILY Daphnidae Daphnia (all species in genus) flea, water ORDER Decapoda FAMILY Atelecyclidae Erimacrus isenbeckii crab, horsehair FAMILY Cancridae Cancer antennarius crab, California rock Cancer anthonyi crab, yellowstone Cancer borealis crab, Jonah Cancer magister crab, dungeness Cancer productus crab, rock (red) FAMILY Geryonidae Geryon affinis crab, golden FAMILY Lithodidae Paralithodes camtschatica crab, Alaskan king FAMILY Majidae Chionocetes bairdi crab, snow Chionocetes opilio crab, snow 1 CONDITIONAL ANIMAL LIST §4-71-6.5 SCIENTIFIC NAME COMMON NAME Chionocetes tanneri crab, snow FAMILY Nephropidae Homarus (all species in genus) lobster, true FAMILY Palaemonidae Macrobrachium lar shrimp, freshwater Macrobrachium rosenbergi prawn, giant long-legged FAMILY Palinuridae Jasus (all species in genus) crayfish, saltwater; lobster Panulirus argus lobster, Atlantic spiny Panulirus longipes femoristriga crayfish, saltwater Panulirus pencillatus lobster, spiny FAMILY Portunidae Callinectes sapidus crab, blue Scylla serrata crab, Samoan; serrate, swimming FAMILY Raninidae Ranina ranina crab, spanner; red frog, Hawaiian CLASS Insecta ORDER Coleoptera FAMILY Tenebrionidae Tenebrio molitor mealworm, -

Hardy Tetra Community Aquarium
Elmer’s Aquarium Community Tank Ideas Tank Size: 10 gal or more H ardy Tetra Community Aquarium Why Keep Them? This community is our most popular community tank for beginners with tanks of 10 to 30 gallons. Tetras represent a large group of fish, and many of them make great choices for a beginner’s community tank. Many tetras are active, colorful, hardy and get along well with other tank mates. Housing: This community tank is suitable for tanks of 10 gallon and up. Filtration can include an AquaClear power filter with a supplemental air pump and sponge filter. They like to swim in the middle water layers. Provide some bushy plants (live or plastic) toward the rear of the tank, and leave the front open for swimming. Water Conditions: Temperature 74-80. pH- (6.6-7.2) Use Seachem Neutral Regulator with each partial water change. Live Plants: Live plants are highly recommended for this community, but not required. Live plants will help bring out the best coloration and behavior in these fish as well as maintain optimal tank conditions. Feeding: Feed this community two to three small feedings per day. Feed flakes, small pellets, and some assorted frozen foods such as mysis shrimp for the most nutrition. How Many? Tetras, Danios, Rasboras, Barbs, Moons and Swordtails are schooling fish and should be bought in groups of at least 3 or more. Tank Mates: Choose other fish of similar size and temperament. Our staff can help you find them in our store. Tetras: Black Skirt Tetra, White Skirt Tetra, Serpae Tetra, Glowlight Tetra, Flame Von Rio Tetra, Bleeding Heart Tetra, Pristella Tetra, Black Phantom Tetra, Red Phantom Tetra, Silver Tip Tetra, Red Eye Tetra, Gold Tetra, Diamond Tetra, Emperor Tetra, GloFish Tetra, Bloodfin Tetra, Head & Tail Light Tetra, Rasboras: Harlequin Rasbora, Brilliant Rasbora, Scissortail Rasbora. -

Blackline Rasbora (Rasbora Borapetensis) ERSS
Blackline Rasbora (Rasbora borapetensis) Ecological Risk Screening Summary U.S. Fish & Wildlife Service, July 2010 Revised, March 2019 Web Version, 9/13/2019 Photo: Lerdsuwa. Licensed under Creative Commons Attribution-Share Alike 3.0 Unported. Available: https://commons.wikimedia.org/wiki/File:Rasbora_borapetensis.jpg. (March 2019). 1 Native Range and Status in the United States Native Range From Froese and Pauly (2019): “Asia: Mekong [Cambodia, Laos, Thailand, Vietnam], Chao Phraya and Meklong basins [Thailand]; also northern Malay Peninsula.” “[In Cambodia:] Found in Stung O Krien, Stung Po Ben, Beng Kebal Damrey, Stung Chihreng, Tuk Sap, Snoc Trou, Spean Tros [Kottelat 1985].” 1 “[In Laos:] Found in the middle Xe Bangfai River [Kottelat 1998], Tha Ngon, Tha Bo, Sithan Tay, Pakse, Hatsalao and Pathoum Phon of the Mekong basin [Taki 1974].” “[In Thailand:] Found in Mekong, Meklong and Chao Phraya basins and in the northern Malay Peninsula [Kottelat 1998]. Collected in the Mekong basin at Nam Man about 2 km upstream of Amphoe Dan Sai in Loei Province [Kottelat 1990]; also from Khon Kaen, Chachoengsao, Sakon Nakhon, Nakhon Sawan, Nakhon Ratchasima, Narathiwat, Phrae, Chanthaburi, Phitsanulok, Prachin Buri and Surat Thani [Monkolprasit et al. 1997].” From Vidthayanon (2012): “Brunei Darussalam; Cambodia; Indonesia (Sumatera, Kalimantan); Lao People's Democratic Republic; Malaysia (Sarawak, Sabah, Peninsular Malaysia); Thailand; Viet Nam” Status in the United States No records of Rasbora borapetensis in the wild in the United States were found. R. borapetensis is in trade in the United States. From That Pet Place (2019): “Brilliant Rasbora – Rasbora borapetensis $1.99” Means of Introductions in the United States No records of Rasbora borapetensis in the wild in the United States were found. -

Parent Mill Mill Name Latitude Longitude Country Aa Sawit Siang
PepsiCo Palm Oil Mill List 2018 The following list is of mills that were in our supply chain in 2018 and does not necessarily reflect mills that are supplying or will supply PepsiCo in 2019. Some of these mills are associated with ongoing complaints that have been registered in our Grievance Mechanism and are being managed through our grievance process. The following palm oil mill list is based on information that has been self-reported to us by suppliers and has only been partially independently verified (see our Palm Oil Progress Report for more information). Though we have made considerable effort to validate the data, we cannot guarantee its full accuracy or completeness. Parent Mill Mill Name Latitude Longitude Country Aa Sawit Siang 1.545386 104.209347 Malaysia Aathi Bagawathi Manufacturing Abdi Budi Mulia 2.051269 100.252339 Indonesia Aathi Bagawathi Manufacturing Abdi Budi Mulia 2 2.11272 100.27311 Indonesia Ace Oil Mill Ace Oil Mill 2.91192 102.77981 Malaysia Aceites Aceites Cimarrones 3.035593889 -73.11146556 Colombia Aceites De Palma Aceites De Palma 18.0470389 -94.91766389 Mexico Aceites Manuelita Yaguarito 3.883139 -73.339917 Colombia Aceites Manuelita Manavire 3.937706 -73.36539 Colombia Aceites Sustentables De Palma Aceites Sustentables De Palma 16.360506 -90.467794 Mexico Achi Jaya Plantations Johor Labis 2.251472222 103.0513056 Malaysia Adimulia Agrolestari Singingi -0.205611 101.318944 Indonesia Adimulia Agrolestari Segati -0.108983 101.386783 Indonesia Adimulia Palmo Lestari Adimulia Palmo Lestari -1.705469 102.867739 -

Mill Name Parent Company Country State Or Province
MILL NAME PARENT COMPANY COUNTRY STATE OR PROVINCE DISTRICT 1 Abago Braganza Colombia Meta Puerto Gaitán 2 Abdi Budi Mulia Aathi Bagawathi Manufacturing Indonesia Sumatera Utara Labuhanbatu Selatan 3 Abedon Kretam Holdings Malaysia Sabah Semporna 4 Ace Oil Mill Ace Oil Mill Malaysia Pahang Rompin 5 Aceitera Chiapaneca Blanca Palomeras Mexico Chiapas Acapetahua 6 Aceites CI Biocosta Colombia Magdalena Aracataca 7 Aceites Cimarrones Aceites Colombia Meta Puerto Rico 8 Aceites De Palma Aceites De Palma Mexico Veracruz Hueyapan de Ocampo 9 Aceites Morichal CI Biocosta Colombia Meta San Carlos de Guaroa 10 Aceites Sustentables De Palma Aceites Sustentables De Palma Mexico Chiapas Ocosingo 11 Aceydesa Aceydesa Honduras Colón Trujillo 12 Adei Plantation Nilo 1 Kuala Lumpur Kepong Indonesia Riau Pelalawan 13 Adei Plantation Nilo 2 Kuala Lumpur Kepong Indonesia Riau Pelalawan 14 Adela Felda Global Ventures Malaysia Johor Kota Tinggi 15 Adimulia Palmo Lestari Adimulia Palmo Lestari Indonesia Jambi Batang Hari 16 Adolina Perkebunan Nusantara IV Indonesia Sumatera Utara Serdang Bedagai 17 Aek Loba Socfin Group Indonesia Sumatera Utara Asahan 18 Aek Nabara Selatan Perkebunan Nusantara III Indonesia Sumatera Utara Labuhanbatu 19 Aek Nopan Kencana Inti Perkasa Indonesia Sumatera Utara Labuhanbatu Utara 20 Aek Raso Perkebunan Nusantara III Indonesia Sumatera Utara Labuhanbatu Selatan 21 Aek Sibirong Maju Indo Raya Indonesia Sumatera Utara Tapanuli Selatan 22 Aek Tinga Mandiri Sawit Bersama Indonesia Sumatera Utara Padang Lawas 23 Aek Torop Perkebunan -

Celestial Pearl Danio", a New Genus and Species of Colourful Minute Cyprinid Fish from Myanmar (Pisces: Cypriniformes)
THE RAFFLES BULLETIN OF ZOOLOGY 2007 55(1): 131-140 Date of Publication: 28 Feb.2007 © National University of Singapore THE "CELESTIAL PEARL DANIO", A NEW GENUS AND SPECIES OF COLOURFUL MINUTE CYPRINID FISH FROM MYANMAR (PISCES: CYPRINIFORMES) Tyson R. Roberts Research Associate, Smithsonian Tropical Research Institute Email: [email protected] ABSTRACT. - Celestichthys margaritatus, a new genus and species of Danioinae, is described from a rapidly developing locality in the Salween basin about 70-80 km northeast of Inle Lake in northern Myanmar. Males and females are strikingly colouful. It is apparently most closely related to two danioins endemic to Inle, Microrasbora rubescens and "Microrasbora" erythromicron. The latter species may be congeneric with the new species. The new genus is identified as a danioin by specializations on its lower jaw and its numerous anal fin rays. The colouration, while highly distinctive, seems also to be characteristically danioin. The danioin notch (Roberts, 1986; Fang, 2003) is reduced or absent, but the danioin mandibular flap and bony knob (defined herein) are present. The anal fin has iiiSVz-lOV: rays. In addition to its distinctive body spots and barred fins the new fish is distinguished from other species of danioins by the following combination of characters: snout and mouth extremely short; premaxillary with an elongate and very slender ascending process; mandible foreshortened; body deep, with rounded dorsal and anal fins; modal vertebral count 15+16=31; caudal fin moderately rather than deeply forked; principal caudal fin rays 9/8; scales vertically ovoid; and pharyngeal teeth conical, in three rows KEY WORDS. - Hopong; principal caudal fin rays; danioin mandibular notch, knob, and pad; captive breeding. -

The AQUATIC DESIGN CENTRE
The AQUATIC DESIGN CENTRE ltd 26 Zennor Road Trade Park, Balham, SW12 0PS Ph: 020 7580 6764 [email protected] PLEASE CALL TO CHECK AVAILABILITY ON DAY Complete Freshwater Livestock (2019) Livebearers Common Name In Stock Y/N Limia melanogaster Y Poecilia latipinna Dalmatian Molly Y Poecilia latipinna Silver Lyre Tail Molly Y Poecilia reticulata Male Guppy Asst Colours Y Poecilia reticulata Red Cap, Cobra, Elephant Ear Guppy Y Poecilia reticulata Female Guppy Y Poecilia sphenops Molly: Black, Canary, Silver, Marble. y Poecilia velifera Sailfin Molly Y Poecilia wingei Endler's Guppy Y Xiphophorus hellerii Swordtail: Pineapple,Red, Green, Black, Lyre Y Xiphophorus hellerii Kohaku Swordtail, Koi, HiFin Xiphophorus maculatus Platy: wagtail,blue,red, sunset, variatus Y Tetras Common Name Aphyocarax paraguayemsis White Tip Tetra Aphyocharax anisitsi Bloodfin Tetra Y Arnoldichthys spilopterus Red Eye Tetra Y Axelrodia riesei Ruby Tetra Bathyaethiops greeni Red Back Congo Tetra Y Boehlkea fredcochui Blue King Tetra Copella meinkeni Spotted Splashing Tetra Crenuchus spilurus Sailfin Characin y Gymnocorymbus ternetzi Black Widow Tetra Y Hasemania nana Silver Tipped Tetra y Hemigrammus erythrozonus Glowlight Tetra y Hemigrammus ocelifer Beacon Tetra y Hemigrammus pulcher Pretty Tetra y Hemigrammus rhodostomus Diamond Back Rummy Nose y Hemigrammus rhodostomus Rummy nose Tetra y Hemigrammus rubrostriatus Hemigrammus vorderwimkieri Platinum Tetra y Hyphessobrycon amandae Ember Tetra y Hyphessobrycon amapaensis Amapa Tetra Y Hyphessobrycon bentosi -
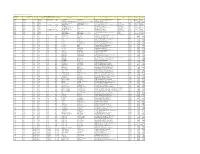
Colgate Palmolive List of Mills As of June 2018 (H1 2018) Direct
Colgate Palmolive List of Mills as of June 2018 (H1 2018) Direct Supplier Second Refiner First Refinery/Aggregator Information Load Port/ Refinery/Aggregator Address Province/ Direct Supplier Supplier Parent Company Refinery/Aggregator Name Mill Company Name Mill Name Country Latitude Longitude Location Location State AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora La Francia Guatemala Extractora Agroaceite Extractora Agroaceite Finca Pensilvania Aldea Los Encuentros, Coatepeque Quetzaltenango. Coatepeque Guatemala 14°33'19.1"N 92°00'20.3"W AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora del Atlantico Guatemala Extractora del Atlantico Extractora del Atlantico km276.5, carretera al Atlantico,Aldea Champona, Morales, izabal Izabal Guatemala 15°35'29.70"N 88°32'40.70"O AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora La Francia Guatemala Extractora La Francia Extractora La Francia km. 243, carretera al Atlantico,Aldea Buena Vista, Morales, izabal Izabal Guatemala 15°28'48.42"N 88°48'6.45" O Oleofinos Oleofinos Mexico Pasternak - - ASOCIACION AGROINDUSTRIAL DE PALMICULTORES DE SABA C.V.Asociacion (ASAPALSA) Agroindustrial de Palmicutores de Saba (ASAPALSA) ALDEA DE ORICA, SABA, COLON Colon HONDURAS 15.54505 -86.180154 Oleofinos Oleofinos Mexico Pasternak - - Cooperativa Agroindustrial de Productores de Palma AceiteraCoopeagropal R.L. (Coopeagropal El Robel R.L.) EL ROBLE, LAUREL, CORREDORES, PUNTARENAS, COSTA RICA Puntarenas Costa Rica 8.4358333 -82.94469444 Oleofinos Oleofinos Mexico Pasternak - - CORPORACIÓN -

Buku Panduan Pertemuan Ilmiah Perhimpunan Biologi Indonesia (PBI) the Indonesian Biological Society Scientific Meeting
Buku Panduan Pertemuan Ilmiah Perhimpunan Biologi Indonesia (PBI) The Indonesian Biological Society Scientific Meeting “Peran biologi dalam pendayagunaan bioresources Indonesia untuk meningkatkan daya saing bangsa The roles of Biology in utilization of Indonesian bioresources to improve nation competitiveness ” Seminar Nasional ke-22 Simposium Internasional Biodiversitas Indonesia International Symposium on Indonesian Biodiversity Indonesian Biological Society Universitas Jenderal Soedirman Purwokerto | 31 Agustus - 1 September 2013 Pertemuan Ilmiah Perhimpunan Biologi Indonesia KATA PENGANTAR Buku panduan ini disusun untuk memudahkan peserta mengikuti Kongres XV, Seminar Nasional XXII, dan Simposium Internasional Biodiversitas Indonesia Perhimpunan Biologi Indonesia yang dilaksanakan pada tanggal 30 Agustus hingga 1 September 2013 di Universitas Jenderal Soedirman Purwokerto dengan tema “Peran biologi dalam pendayagunaan bioresources Indonesia untuk meningkatkan daya saing bangsa”. Buku panduan ini memuat jadwal acara dan seluruh abstrak makalah yang disajikan dalam pertemuan ilmiah PBI tahun 2013. Makalah dikelompokkan dalam enam topik (simposia) yaitu: (1) Pendidikan Biologi; (2) Biologi Sel, Biologi Molekuler, dan Genetika; (3) Fisiologi dan Reproduksi; (4) Ekologi dan Konservasi; dan (5) Biologi Umum yang merupakan Simposia Nasional; serta (6) Biodiversitas Indonesia yang merupakan Simposium Internasional. Makalah disajikan dengan dua kategori yaitu oral dan poster. Makalah yang diajukan untuk diterbitkan dalam Prosiding Seminar Nasional -

Mill List - 2020
General Mills - Mill List - 2020 General Mills July 2020 - December 2020 Parent Mill Name Latitude Longitude RSPO Country State or Province District UML ID 3F Oil Palm Agrotech 3F Oil Palm Agrotech 17.00352 81.46973 No India Andhra Pradesh West Godavari PO1000008590 Aathi Bagawathi Manufacturing Abdi Budi Mulia 2.051269 100.252339 No Indonesia Sumatera Utara Labuhanbatu Selatan PO1000004269 Aathi Bagawathi Manufacturing Abdi Budi Mulia 2 2.11272 100.27311 No Indonesia Sumatera Utara Labuhanbatu Selatan PO1000008154 Abago Extractora Braganza 4.286556 -72.134083 No Colombia Meta Puerto Gaitán PO1000008347 Ace Oil Mill Ace Oil Mill 2.91192 102.77981 No Malaysia Pahang Rompin PO1000003712 Aceites De Palma Aceites De Palma 18.0470389 -94.91766389 No Mexico Veracruz Hueyapan de Ocampo PO1000004765 Aceites Morichal Aceites Morichal 3.92985 -73.242775 No Colombia Meta San Carlos de Guaroa PO1000003988 Aceites Sustentables De Palma Aceites Sustentables De Palma 16.360506 -90.467794 No Mexico Chiapas Ocosingo PO1000008341 Achi Jaya Plantations Johor Labis 2.251472222 103.0513056 No Malaysia Johor Segamat PO1000003713 Adimulia Agrolestari Segati -0.108983 101.386783 No Indonesia Riau Kampar PO1000004351 Adimulia Agrolestari Surya Agrolika Reksa (Sei Basau) -0.136967 101.3908 No Indonesia Riau Kuantan Singingi PO1000004358 Adimulia Agrolestari Surya Agrolika Reksa (Singingi) -0.205611 101.318944 No Indonesia Riau Kuantan Singingi PO1000007629 ADIMULIA AGROLESTARI SEI TESO 0.11065 101.38678 NO INDONESIA Adimulia Palmo Lestari Adimulia Palmo Lestari -

AACL Bioflux, Volume 9(4) August 30, 2016
AACL Bioflux, Volume 9(4) August 30, 2016 Contents Hamsiah, Herawati E. Y., Mahmudi M., Sartimbul A., 2016 Seasonal variation of bivalve diversity in seagrass ecosystem of Labakkang coastal water, Pangkep, South Sulawesi, Indonesia. AACL Bioflux 9(4):775-784. Purnama A. A., Yolanda R., 2016 Diversity of freshwater fish (Pisces) in Kumu River, Rokan Hulu District, Riau Province, Indonesia. AACL Bioflux 9(4):785-789. Sriwulan, Anshary H., 2016 Occurrence of infectious and non infectious Decapod Penstyldensovirus 1 (PstDV-1) from tiger shrimp (Penaeus monodon) in South Sulawesi, Indonesia. AACL Bioflux 9(4):790-801. Rizal M., Wiryawan B., Wisudo S. H., Solihin I., Haluan J., 2016 Institutional development strategy through Interpretive Structural Modelling (ISM) for gillnet fisher group in Barsela Aceh, Indonesia. AACL Bioflux 9(4):802-814. Putro S. P., Widowati, Rahmansyah R., Suminto S., 2016 Application of polyculture using stratified double net cage: a case study at Awerange Gulf, Barru, South Sulawesi, Indonesia. AACL Bioflux 9(4):815-822. Effendi I., Suprayudi M. A., Surawidjaja E. H., Supriyono E., Zairin M. Jr., Sukenda, 2016 Production performance of white shrimp (Litopenaeus vannamei) under sea floating net cages with biofloc and periphyton juvenile. AACL Bioflux 9(4):823-832. Yudha I. G., Rahardjo M. F., Djokosetiyanto D., Batu D. T. F. L., 2016 Some aspects of the reproductive biology of Labiobarbus ocellatus in Tulang Bawang River, Lampung, Indonesia. AACL Bioflux 9(4):833-844. Manan H., Moh J. H. Z., Kasan N. A., Suratman S., Ikhwanuddin M., 2016 Study on carbon sinks by classified biofloc phytoplankton from marine shrimp pond water. -

BMC Evolutionary Biology Biomed Central
BMC Evolutionary Biology BioMed Central Research article Open Access Evolution of miniaturization and the phylogenetic position of Paedocypris, comprising the world's smallest vertebrate Lukas Rüber*1, Maurice Kottelat2, Heok Hui Tan3, Peter KL Ng3 and Ralf Britz1 Address: 1Department of Zoology, The Natural History Museum, Cromwell Road, London SW7 5BD, UK, 2Route de la Baroche 12, Case postale 57, CH-2952 Cornol, Switzerland (permanent address) and Raffles Museum of Biodiversity Research, National University of Singapore, Kent Ridge, Singapore 119260 and 3Department of Biological Sciences, National University of Singapore, Kent Ridge, Singapore 119260 Email: Lukas Rüber* - [email protected]; Maurice Kottelat - [email protected]; Heok Hui Tan - [email protected]; Peter KL Ng - [email protected]; Ralf Britz - [email protected] * Corresponding author Published: 13 March 2007 Received: 23 October 2006 Accepted: 13 March 2007 BMC Evolutionary Biology 2007, 7:38 doi:10.1186/1471-2148-7-38 This article is available from: http://www.biomedcentral.com/1471-2148/7/38 © 2007 Rüber et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Paedocypris, a highly developmentally truncated fish from peat swamp forests in Southeast Asia, comprises the world's smallest vertebrate. Although clearly a cyprinid fish, a hypothesis about its phylogenetic position among the subfamilies of this largest teleost family, with over 2400 species, does not exist.