Biology, Breeding and Applications of Cannabis Sativa
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FLORA from FĂRĂGĂU AREA (MUREŞ COUNTY) AS POTENTIAL SOURCE of MEDICINAL PLANTS Silvia OROIAN1*, Mihaela SĂMĂRGHIŢAN2
ISSN: 2601 – 6141, ISSN-L: 2601 – 6141 Acta Biologica Marisiensis 2018, 1(1): 60-70 ORIGINAL PAPER FLORA FROM FĂRĂGĂU AREA (MUREŞ COUNTY) AS POTENTIAL SOURCE OF MEDICINAL PLANTS Silvia OROIAN1*, Mihaela SĂMĂRGHIŢAN2 1Department of Pharmaceutical Botany, University of Medicine and Pharmacy of Tîrgu Mureş, Romania 2Mureş County Museum, Department of Natural Sciences, Tîrgu Mureş, Romania *Correspondence: Silvia OROIAN [email protected] Received: 2 July 2018; Accepted: 9 July 2018; Published: 15 July 2018 Abstract The aim of this study was to identify a potential source of medicinal plant from Transylvanian Plain. Also, the paper provides information about the hayfields floral richness, a great scientific value for Romania and Europe. The study of the flora was carried out in several stages: 2005-2008, 2013, 2017-2018. In the studied area, 397 taxa were identified, distributed in 82 families with therapeutic potential, represented by 164 medical taxa, 37 of them being in the European Pharmacopoeia 8.5. The study reveals that most plants contain: volatile oils (13.41%), tannins (12.19%), flavonoids (9.75%), mucilages (8.53%) etc. This plants can be used in the treatment of various human disorders: disorders of the digestive system, respiratory system, skin disorders, muscular and skeletal systems, genitourinary system, in gynaecological disorders, cardiovascular, and central nervous sistem disorders. In the study plants protected by law at European and national level were identified: Echium maculatum, Cephalaria radiata, Crambe tataria, Narcissus poeticus ssp. radiiflorus, Salvia nutans, Iris aphylla, Orchis morio, Orchis tridentata, Adonis vernalis, Dictamnus albus, Hammarbya paludosa etc. Keywords: Fărăgău, medicinal plants, human disease, Mureş County 1. -

TAXON:Trema Orientalis (L.) Blume SCORE:10.0 RATING
TAXON: Trema orientalis (L.) Blume SCORE: 10.0 RATING: High Risk Taxon: Trema orientalis (L.) Blume Family: Cannabaceae Common Name(s): charcoal tree Synonym(s): Celtis guineensis Schumach. gunpowder tree Celtis orientalis L. peach cedar Trema guineensis (Schumach.) Ficalho poison peach Assessor: Chuck Chimera Status: Assessor Approved End Date: 4 Mar 2020 WRA Score: 10.0 Designation: H(Hawai'i) Rating: High Risk Keywords: Tropical, Pioneer Tree, Weedy, Bird-Dispersed, Coppices Qsn # Question Answer Option Answer 101 Is the species highly domesticated? y=-3, n=0 n 102 Has the species become naturalized where grown? 103 Does the species have weedy races? Species suited to tropical or subtropical climate(s) - If 201 island is primarily wet habitat, then substitute "wet (0-low; 1-intermediate; 2-high) (See Appendix 2) High tropical" for "tropical or subtropical" 202 Quality of climate match data (0-low; 1-intermediate; 2-high) (See Appendix 2) High 203 Broad climate suitability (environmental versatility) y=1, n=0 y Native or naturalized in regions with tropical or 204 y=1, n=0 y subtropical climates Does the species have a history of repeated introductions 205 y=-2, ?=-1, n=0 y outside its natural range? 301 Naturalized beyond native range y = 1*multiplier (see Appendix 2), n= question 205 y 302 Garden/amenity/disturbance weed n=0, y = 1*multiplier (see Appendix 2) y 303 Agricultural/forestry/horticultural weed n=0, y = 2*multiplier (see Appendix 2) y 304 Environmental weed n=0, y = 2*multiplier (see Appendix 2) n 305 Congeneric weed 401 -

Familial Mediterranean Fever
:: Familial Mediterranean fever – This document is a translation of the French recommendations drafted by Prof Gilles Grateau, Dr Véronique Hentgen, Dr Katia Stankovic Stojanovic and Dr Gilles Bagou reviewed and published by Orphanet in 2010. – Some of the procedures mentioned, particularly drug treatments, may not be validated in the country where you practice. Synonyms: Periodic disease, FMF Definition: Familial Mediterranean fever (FMF) is an auto-inflammatory disease of genetic origin, affecting Mediterranean populations and characterised by recurrent attacks of fever accompanied by polyserositis that causes the symptoms. Colchicine is the basic reference treatment and is designed to tackle inflammatory attacks and prevent amyloidosis, the most severe complication of FMF. Further information: See the Orphanet abstract http://www.orpha.net/data/patho/Pro/en/Emergency_FamilialMediterraneanFever-enPro920.pdf ©Orphanet UK 1/5 Pre-hospital emergency care recommendations Call for a patient suffering from Familial mediterranean fever Synonyms periodic disease FMF Mechanisms auto-inflammatory disease that affects Mediterranean populations in particular, due to mutation of the MEFV gene that codes pyrin or marenostrin, this being the underlying cause of congenital immune dysfunction; repeated inflammatory attacks can lead to amyloidosis, particularly renal Specific risks in emergency situations acute inflammatory attack, particularly abdominal (pseudo-surgical), but also thoracic, articular (knees) or testicular fever as an expression -

Cannabaceae Studies Two Day Virtual Conference at COS
Siskiyou Joint Community College District 800 College Avenue Weed, CA 96094 Telephone: (530) 938-5555 Toll-Free: (888) 397-4339 PRESS RELEASE Public Relations Office: FOR IMMEDIATE RELEASE Dawnie Slabaugh, Director: (530) 938-5373 ReleaseRelease #: 029 57 Email: [email protected] Release #: 04455 Date: OctoberOctober 23,23, 20152015 Date:Date: AugustMarch 2611, 20172021 Cannabaceae Studies Two Day Virtual Conference at COS Weed / COS – College of the Siskiyous is proud to announce, the launch of its Cannabaceae Studies program through a two-day Cannabaceae Studies Virtual Conference scheduled for April 17 and April 18. During the virtual conference, registered participants will complete four Cannabaceae Studies Community Education courses taught by experts in the field: • Science of the Cannabaceae Family: Overview of the peer-reviewed scientific literature relevant to the therapeutic efficacy and relative safety of Cannabaceae plants including hemp and cannabis. Instructor: Nick Joslin – Lemon Remedy • Federal v. State Law: Introduction to the legal issues surrounding medical cannabis and the hemp/cannabis industry. Instructor: Nicole Howell, Esq. – Clark Howell LLP Lawyers for the Green Frontier • Politics v. History: Learn about the political history of cannabis, and the key players and policies that have shaped the policy as we know it today. Instructor: Raymond Strack – CEO Shasta- Jefferson Growth Business • Industrial Hemp Essentials: Learn about the basic farming practices for the different applications, of the respective parts of the hemp plant. Instructor: Tyler Hoff – Founder Bulk Hemp Warehouse The cost to participate is $50 for the full program or $15 per class. Don’t miss this unique opportunity to get hempeducated! To register visit - https://college-of-the-siskiyous.coursestorm.com. -

On the Origin of Hops: Genetic Variability, Phylogenetic Relationships, and Ecological Plasticity of Humulus (Cannabaceae)
ON THE ORIGIN OF HOPS: GENETIC VARIABILITY, PHYLOGENETIC RELATIONSHIPS, AND ECOLOGICAL PLASTICITY OF HUMULUS (CANNABACEAE) A DISSERTATION SUBMITTED TO THE GRADUATE DIVISION OF THE UNIVERSITY OF HAWAI‘I AT MĀNOA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY IN BOTANY MAY 2014 By Jeffrey R. Boutain DISSERTATION COMMITTEE: Will C. McClatchey, Chairperson Mark D. Merlin Sterling C. Keeley Clifford W. Morden Stacy Jørgensen Copyright © 2014 by Jeffrey R. Boutain ii This dissertation is dedicated to my family tree. iii ACKNOWLEDGEMENTS There are a number of individuals to whom I am indebted in many customs. First and foremost, I thank my committee members for their contribution, patience, persistence, and motivation that helped me complete this dissertation. Specifically, thank you Dr. Will McClatchey for the opportunity to study in a botany program with you as my advisor and especially the encouragement to surf plant genomes. Also with great gratitude, thank you Dr. Sterling Keeley for the opportunity to work on much of this dissertation in your molecular phylogenetics and systematics lab. In addition, thank you Dr. Mark Merlin for numerous brainstorming sessions as well as your guidance and expert perspective on the Cannabaceae. Also, thank you Dr. Cliff Morden for the opportunity to work in your lab where the beginnings of this molecular research took place. Thank you Dr. Jianchu Xu for welcoming me into your lab group at the Kunming Institute of Botany, Chinese Academy of Sciences (CAS) and the opportunity to study the Yunnan hop. In many ways, major contributions towards the completion of this dissertation have come from my family, and I thank you for your unconditional encouragement, love, and support. -

Guideline for Preoperative Medication Management
Guideline: Preoperative Medication Management Guideline for Preoperative Medication Management Purpose of Guideline: To provide guidance to physicians, advanced practice providers (APPs), pharmacists, and nurses regarding medication management in the preoperative setting. Background: Appropriate perioperative medication management is essential to ensure positive surgical outcomes and prevent medication misadventures.1 Results from a prospective analysis of 1,025 patients admitted to a general surgical unit concluded that patients on at least one medication for a chronic disease are 2.7 times more likely to experience surgical complications compared with those not taking any medications. As the aging population requires more medication use and the availability of various nonprescription medications continues to increase, so does the risk of polypharmacy and the need for perioperative medication guidance.2 There are no well-designed trials to support evidence-based recommendations for perioperative medication management; however, general principles and best practice approaches are available. General considerations for perioperative medication management include a thorough medication history, understanding of the medication pharmacokinetics and potential for withdrawal symptoms, understanding the risks associated with the surgical procedure and the risks of medication discontinuation based on the intended indication. Clinical judgement must be exercised, especially if medication pharmacokinetics are not predictable or there are significant risks associated with inappropriate medication withdrawal (eg, tolerance) or continuation (eg, postsurgical infection).2 Clinical Assessment: Prior to instructing the patient on preoperative medication management, completion of a thorough medication history is recommended – including all information on prescription medications, over-the-counter medications, “as needed” medications, vitamins, supplements, and herbal medications. Allergies should also be verified and documented. -

Contribution to the Biosystematics of Celtis L. (Celtidaceae) with Special Emphasis on the African Species
Contribution to the biosystematics of Celtis L. (Celtidaceae) with special emphasis on the African species Ali Sattarian I Promotor: Prof. Dr. Ir. L.J.G. van der Maesen Hoogleraar Plantentaxonomie Wageningen Universiteit Co-promotor Dr. F.T. Bakker Universitair Docent, leerstoelgroep Biosystematiek Wageningen Universiteit Overige leden: Prof. Dr. E. Robbrecht, Universiteit van Antwerpen en Nationale Plantentuin, Meise, België Prof. Dr. E. Smets Universiteit Leiden Prof. Dr. L.H.W. van der Plas Wageningen Universiteit Prof. Dr. A.M. Cleef Wageningen Universiteit Dr. Ir. R.H.M.J. Lemmens Plant Resources of Tropical Africa, WUR Dit onderzoek is uitgevoerd binnen de onderzoekschool Biodiversiteit. II Contribution to the biosystematics of Celtis L. (Celtidaceae) with special emphasis on the African species Ali Sattarian Proefschrift ter verkrijging van de graad van doctor op gezag van rector magnificus van Wageningen Universiteit Prof. Dr. M.J. Kropff in het openbaar te verdedigen op maandag 26 juni 2006 des namiddags te 16.00 uur in de Aula III Sattarian, A. (2006) PhD thesis Wageningen University, Wageningen ISBN 90-8504-445-6 Key words: Taxonomy of Celti s, morphology, micromorphology, phylogeny, molecular systematics, Ulmaceae and Celtidaceae, revision of African Celtis This study was carried out at the NHN-Wageningen, Biosystematics Group, (Generaal Foulkesweg 37, 6700 ED Wageningen), Department of Plant Sciences, Wageningen University, the Netherlands. IV To my parents my wife (Forogh) and my children (Mohammad Reza, Mobina) V VI Contents ——————————— Chapter 1 - General Introduction ....................................................................................................... 1 Chapter 2 - Evolutionary Relationships of Celtidaceae ..................................................................... 7 R. VAN VELZEN; F.T. BAKKER; A. SATTARIAN & L.J.G. VAN DER MAESEN Chapter 3 - Phylogenetic Relationships of African Celtis (Celtidaceae) ........................................ -

The Effect of Caffeine and Trifluralin on Chromosome Doubling in Wheat
plants Article The Effect of Caffeine and Trifluralin on Chromosome Doubling in Wheat Anther Culture Sue Broughton *, Marieclaire Castello, Li Liu, Julie Killen, Anna Hepworth and Rebecca O’Leary Department of Primary Industries and Regional Development, 3 Baron-Hay Court, South Perth 6151, Western Australia, Australia; [email protected] (M.C.); [email protected] (L.L.); [email protected] (J.K.); [email protected] (A.H.); Rebecca.O'[email protected] (R.O.) * Correspondence: [email protected] Received: 3 December 2019; Accepted: 10 January 2020; Published: 15 January 2020 Abstract: Challenges for wheat doubled haploid (DH) production using anther culture include genotype variability in green plant regeneration and spontaneous chromosome doubling. The frequency of chromosome doubling in our program can vary from 14% to 80%. Caffeine or trifluralin was applied at the start of the induction phase to improve early genome doubling. Caffeine treatment at 0.5 mM for 24 h significantly improved green plant production in two of the six spring wheat crosses but had no effect on the other crosses. The improvements were observed in Trojan/Havoc and Lancer/LPB14-0392, where green plant numbers increased by 14% and 27% to 161 and 42 green plants per 30 anthers, respectively. Caffeine had no significant effect on chromosome doubling, despite a higher frequency of doubling in several caffeine treatments in the first experiment (67–68%) compared to the control (56%). In contrast, trifluralin significantly improved doubling following a 48 h treatment, from 38% in the control to 51% and 53% in the 1 µM and 3 µM trifluralin treatments, respectively. -

Your 2017 Three-Tier
Your 2017 Three-Tier Prescription Drug List effective January 1, 2017 Connecticut Advantage Three-Tier Please read: This document contains information about commonly prescribed medications. This Prescription Drug List (PDL) is accurate as of January 1, 2017 and is subject to change after this date. The next anticipated update will be in July 2017. Your estimated coverage and copay/co-insurance may vary based on the benefit plan you choose and the effective date of the plan. For additional information: Call the toll-free member phone number on your health plan ID card. Visit myuhc.com® • Locate a participating retail pharmacy by ZIP code. • Look up possible lower-cost medication alternatives. • Compare medication pricing and options. 1 Your Prescription Drug List This Prescription Drug List (PDL) outlines covered medications for certain conditions and organizes them into cost levels, also known as tiers. An important part of the PDL is giving you choices so you and your doctor can choose the best course of treatment for you. Go to myuhc.com® for complete drug information Since the PDL may change, we encourage you to visit our website, myuhc.com. This website is the best source for up-to-date information about the medications your pharmacy benefit covers, possible lower-cost options, and cost comparisons. To view PDL information without logging on to the member portal, visit myuhc.com and select “Pharmacy Information” under “Links and Tools.” 2 At UnitedHealthcare, we want to help you better understand your medication options. Your pharmacy benefit offers flexibility and choice in determining the right medication for you. -

Common Names and Latin Synonyms for Humulus Lupulus Sex Female
COMMON NAMES AND LATIN SYNONYMS FOR HUMULUS LUPULUS SEX FEMALE BETIGUERA BINE HOBLON HOMBRECILLO HOP HOP VINE HOPFENS HOPS HOUBLOW LUPIO LUPULO RAZAK VIDARRIA Page 1 of 128 (7/15/2008) ETHNOMEDICAL INFORMATION ON HUMULUS LUPULUS HUMULUS LUPULUS (CANNABACEAE) DRIED AERIAL PARTS ENGLAND USED TO INDUCE SLEEP. STUFFED INTO PILLOWS. PLANT * INHALATION * HUMAN ADULT * T09858 HUMULUS LUPULUS (CANNABACEAE) DRIED BUDS ITALY USED FOR GASTRONOMIC PURPOSES. BUDS * ORAL * HUMAN ADULT * T16715 HUMULUS LUPULUS (CANNABACEAE) ENTIRE PLANT CHINA USED AS A SEDATIVE. DECOCTION * ORAL * HUMAN ADULT * K29113 USED AS A MILD HYPNOTIC. DECOCTION * ORAL * HUMAN ADULT * K29113 USED FOR TUBERCULOSIS. DECOCTION * ORAL * HUMAN ADULT * K29113 USED FOR CANCER. DECOCTION * ORAL * HUMAN ADULT * K29113 USED FOR EXTERNAL ULCERS. DECOCTION * EXTERNAL * HUMAN ADULT * K29113 USED AS AN APHRODISIAC. DECOCTION * ORAL * HUMAN ADULT * MALE * K29113 HUMULUS LUPULUS (CANNABACEAE) ENTIRE PLANT USA USED AS A NERVINE HOT H2O EXT * ORAL * HUMAN ADULT * T01253 Page 2 of 128 (7/15/2008) ETHNOMEDICAL INFORMATION ON HUMULUS LUPULUS HUMULUS LUPULUS (CANNABACEAE) ENTIRE PLANT USA USED FOR DYSMENORRHEA HOT H2O EXT * ORAL * HUMAN ADULT * FEMALE * T01253 HUMULUS LUPULUS (CANNABACEAE) FLOWERING TOPS ITALY USED FOR FRAIL AND LYMPHATIC CHILDREN. INFUSION * ORAL * HUMAN ADULT * T16136 HUMULUS LUPULUS (CANNABACEAE) FLOWERS USA USED AS A SEDATIVE HOT H2O EXT * ORAL * HUMAN ADULT * L00715 HUMULUS LUPULUS (CANNABACEAE) FRUIT USA USED AS A SEDATIVE PLANT * SMOKING * HUMAN ADULT * K04641 HUMULUS LUPULUS (CANNABACEAE) DRIED FRUIT EUROPE USED TO PROMOTE SLEEP. H2O EXT * ORAL * HUMAN ADULT * N12648 HUMULUS LUPULUS (CANNABACEAE) DRIED FRUIT IRAN USED AS AN ANTIPYRETIC. INFUSION * ORAL * HUMAN ADULT * I00004 APPLIED IN JOINT PAIN. INFUSION * EXTERNAL * HUMAN ADULT * I00004 USED AS A HYPNOTIC. -
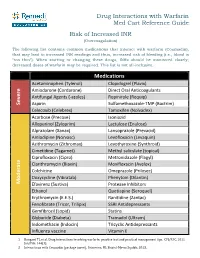
Drug Interactions with Warfarin Med Cart Reference Guide Risk of Increased INR (Overcoagulation)
Drug Interactions with Warfarin Med Cart Reference Guide Risk of Increased INR (Overcoagulation) The following list contains common medications that interact with warfarin (Coumadin), that may lead to increased INR readings and thus, increased risk of bleeding (i.e., blood is “too thin”). When starting or changing these drugs, INRs should be monitored closely; decreased doses of warfarin may be required. This list is not all-inclusive. Medications Acetaminophen (Tylenol) Clopidogrel (Plavix) Amiodarone (Cordarone) Direct Oral Anticoagulants Antifungal Agents (‐azoles) Ropinirole (Requip) Severe Aspirin Sulfamethoxazole‐TMP (Bactrim) Celecoxib (Celebrex) Tamoxifen (Nolvadex) Acarbose (Precose) Isoniazid Allopurinol (Zyloprim) Lactulose (Enulose) Alprazolam (Xanax) Lansoprazole (Prevacid) Amlodipine (Norvasc) Levofloxacin (Levaquin) Azithromycin (Zithromax) Levothyroxine (Synthroid) Cimetidine (Tagamet) Methyl salicylate (topical) Ciprofloxacin (Cipro) Metronidazole (Flagyl) Clarithromycin (Biaxin) Moxifloxacin (Avelox) Colchicine Omeprazole (Prilosec) Doxycycline (Vibratab) Phenytoin (Dilantin) Moderate Efavirenz (Sustiva) Protease Inhibitors Ethanol Quetiapine (Seroquel) Erythromycin (E.E.S.) Ranitidine (Zantac) Fenofibrate (Tricor, Trilipix) SSRI Antidepressants Gemfibrozil (Lopid) Statins Glyburide (Diabeta) Tramadol (Ultram) Indomethacin (Indocin) Tricyclic Antidepressants Influenza vaccine Vitamin E 1. Bungard TJ, et al. Drug interactions involving warfarin: practice tool and practical management tips. CPJ/RPC. 2011 Jan/Feb. 144(1). 2. Interactions with Coumadin [package insert]. Princeton, NJ: Bristol‐Myers Squibb; 2013. Drug Interactions with Warfarin Med Cart Reference Guide Risk of Decreased INR (Undercoagulation) The following list contains common medications that interact with warfarin (Coumadin), which may lead to decreased INR readings and thus, increased risk of clotting, strokes, and DVTs (i.e., blood is “not thin enough”). When starting or changing these drugs, INRs should be monitored closely; increased doses of warfarin may be required. -

An Allergopalynologic Comparative Study of the Humulus Lupulus’ Pollen Grains (Cannabaceae) in Different Habitats
NIKOLETA KALLAJXHIU1, GËZIM KAPIDANI2, PEÇI NAQELLARI1, BLERINA PUPULEKU1, SILVANA TURKU1, ANXHELA DAUTI1, ADMIR JANÇE1 1 Departament of Biology, FNS, “A. Xhuvani” University, Elbasan, Albania, 3001, 2 Departament of Biology, FNS, University of Tirana, Tirana, Albania, 1001 email: [email protected] AN ALLERGOPALYNOLOGIC COMPARATIVE STUDY OF THE HUMULUS LUPULUS’ POLLEN GRAINS (CANNABACEAE) IN DIFFERENT HABITATS ABSTRACT Pollen allergy is a human disease that is rapidly growing among the Albanian population. There are many plants in Albania that cause this disease and Humulus lupulus (Cannabaceae) is one of them. In this study is presented an allergopalyno- logic description of pollen grains of this plant. They were taken in the Elbasan region of Albania and are studied for the first time by the local researchers. The slides were prepared using two methods: acetolysis and basic fuchsine method. The results have shown that the pollen grains of H. lupulus were triporate. Also, is done the comparison of the size of pollen grains for the specie studied with oth- ers from foreign literature that belong to different habitats. The study showed the pollen grains of H. lupulus taken in Elbasan area have similarities to the morpho- logical features with H. lupulus’ pollen according to the foreign literature, but they differ from their size. By our study, it turns out that the pollen grains of H. hupulus from Elbasan habitat have smaller sizes than those obtained from the literature. We conclude that the change in the size of pollen grains caused by environmental factors and the chosen method during processing laboratory. Keywords: Humulus lupulus, allergopalinology, pollen grains, exine, pori.