A Comprehensive Map of Disease Networks and Molecular Drug Discoveries for Glaucoma Haixin Wang1,2,3, Yanhui Deng1, Ling Wan4 & Lulin Huang1,2,3 ✉
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Multi-Drug Rapid Test Panel with Adulteration (Urine)
6-mono-aceto-morphine in urine is 3-7 days. Multi-Drug Rapid Test Panel with 6-MAM 10 Adulteration (Urine) (6-MAM10) The Multi-Drug Rapid Test Panel yields a positive result when the concentration of (±) 3,4-Methylenedioxy- (±) 3,4-Methylenedioxy- benzodiazepines in urine exceeds detective level. Package Insert 500 Buprenorphine (BUP) Amphetamine(MDA500) Amphetamine Instruction Sheet for testing of any combination of the following drugs: Ethyl- β-D-Glucuronide(ETG500) Ethyl- β -D-Glucuronide 500 Buprenorphine is a potent analgesic often used in the treatment of opioid addiction. The drug is ACE/AMP/BAR/BZO/BUP/COC/THC/MTD/MET/MDMA/MOP/MQL/OPI/PCP/PPX/TCA/TML/K sold under the trade names Subutex™, Buprenex™, Temgesic™ and Suboxone™, which ET/OXY/COT/EDDP/FYL/K2/6-MAM/MDA/ETG/CLO/LSD/MPD/ZOL Ethyl- β-D-Glucuronide(ETG1,000) Ethyl- β -D-Glucuronide 1,000 contain Buprenorphine HCl alone or in combination with Naloxone HCl. Therapeutically, Including Specimen Validity Tests (S.V.T.) for: Clonazepam(CLO 400) Clonazepam 400 Buprenorphine is used as a substitution treatment for opioid addicts. Substitution treatment is a Oxidants/PCC, Specific Gravity, pH, Nitrite, Glutaraldehyde and Creatinine Clonazepam(CLO 150) Clonazepam 150 form of medical care offered to opiate addicts (primarily heroin addicts) based on a similar or A rapid test for the simultaneous, qualitative detection of multiple drugs and drug metabolites in identical substance to the drug normally used. In substitution therapy, Buprenorphine is as human urine. For healthcare professionals including professionals at point of care sites. Lysergic Acid Diethylamide (LSD) Lysergic Acid Diethylamide 20 effective as Methadone but demonstrates a lower level of physical dependence. -

Intraocular Pressure Rise After Phacoemulsification Prophylactic
British Journal of Ophthalmology 1995; 79: 809-813 809 Intraocular pressure rise after phacoemulsification Br J Ophthalmol: first published as 10.1136/bjo.79.9.809 on 1 September 1995. Downloaded from with posterior chamber lens implantation: effect of prophylactic medication, wound closure, and surgeon's experience Thomas G Bomer, Wolf-Dietrich A Lagreze, Jens Funk Abstract pressure rises usually occur between 6 and 8 Aims-A prospective clinical trial was hours after surgery.6 carried out to evaluate the effect of Various antiglaucomatous agents have been prophylactic medication, the technique of used to prevent the intraocular pressure rise wound closure, and the surgeon's experi- after cataract extraction. Oral acetazolamide15 16 ence on the intraocular pressure rise after and topical timolol'6-19 lowered the pressure cataract extraction. rise in the early period after intracapsular and Methods-In 100 eyes, the intraocular extracapsular cataract extraction. Levobunolol pressure was measured before as well as proved to be superior to timolol 4-7 hours 2-4, 5-7, and 22-24 hours after phaco- after extracapsular cataract extraction.20 Apra- emulsification and posterior chamber lens clonidine lowered the intraocular pressure rise implantation. Each of 25 patients received after uncomplicated phacoemulsification21 and either 1% topical apraclonidine, 0.5%/o extracapsular cataract extraction22 23 when topical levobunolol, 500 mg oral acetazo- given 30 minutes to 1 hour before surgery, lamide, or placebo. Forty four eyes were whereas immediate postoperative treatment operated with sclerocorneal sutureless with apraclonidine was ineffective.22 24 Miotics tunnel and 56 eyes with corneoscleral are frequently used to promote miosis and incision and suture. -

Product List March 2019 - Page 1 of 53
Wessex has been sourcing and supplying active substances to medicine manufacturers since its incorporation in 1994. We supply from known, trusted partners working to full cGMP and with full regulatory support. Please contact us for details of the following products. Product CAS No. ( R)-2-Methyl-CBS-oxazaborolidine 112022-83-0 (-) (1R) Menthyl Chloroformate 14602-86-9 (+)-Sotalol Hydrochloride 959-24-0 (2R)-2-[(4-Ethyl-2, 3-dioxopiperazinyl) carbonylamino]-2-phenylacetic 63422-71-9 acid (2R)-2-[(4-Ethyl-2-3-dioxopiperazinyl) carbonylamino]-2-(4- 62893-24-7 hydroxyphenyl) acetic acid (r)-(+)-α-Lipoic Acid 1200-22-2 (S)-1-(2-Chloroacetyl) pyrrolidine-2-carbonitrile 207557-35-5 1,1'-Carbonyl diimidazole 530-62-1 1,3-Cyclohexanedione 504-02-9 1-[2-amino-1-(4-methoxyphenyl) ethyl] cyclohexanol acetate 839705-03-2 1-[2-Amino-1-(4-methoxyphenyl) ethyl] cyclohexanol Hydrochloride 130198-05-9 1-[Cyano-(4-methoxyphenyl) methyl] cyclohexanol 93413-76-4 1-Chloroethyl-4-nitrophenyl carbonate 101623-69-2 2-(2-Aminothiazol-4-yl) acetic acid Hydrochloride 66659-20-9 2-(4-Nitrophenyl)ethanamine Hydrochloride 29968-78-3 2,4 Dichlorobenzyl Alcohol (2,4 DCBA) 1777-82-8 2,6-Dichlorophenol 87-65-0 2.6 Diamino Pyridine 136-40-3 2-Aminoheptane Sulfate 6411-75-2 2-Ethylhexanoyl Chloride 760-67-8 2-Ethylhexyl Chloroformate 24468-13-1 2-Isopropyl-4-(N-methylaminomethyl) thiazole Hydrochloride 908591-25-3 4,4,4-Trifluoro-1-(4-methylphenyl)-1,3-butane dione 720-94-5 4,5,6,7-Tetrahydrothieno[3,2,c] pyridine Hydrochloride 28783-41-7 4-Chloro-N-methyl-piperidine 5570-77-4 -
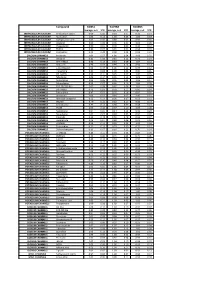
Biomol Average and SD Table S1.Xlsx
Compound GliNS1 G179NS G166NS average, n=5 STD average, n=3 STD average, n=3 STD INTRACELLULAR CALCIUM Antibiotic A-23187 0.00 0.00 0.10 0.07 0.15 0.14 INTRACELLULAR CALCIUM Ryanodine 1.04 0.14 1.03 0.03 1.03 0.03 INTRACELLULAR CALCIUM Cyclopiazonic acid 1.01 0.06 0.88 0.05 0.92 0.06 INTRACELLULAR CALCIUM Gingerol 1.00 0.06 0.91 0.01 1.01 0.06 INTRACELLULAR CALCIUM Thapsigargin 0.00 0.01 0.00 0.00 0.10 0.12 INTRACELLULAR CALCIUM TMB-8 0.89 0.07 0.91 0.05 0.94 0.03 INTRACELLULAR CALCIUM Dantrolene 0.91 0.08 0.98 0.05 0.94 0.01 CALCIUM CHANNELS Amiloride 1.01 0.07 1.01 0.04 1.03 0.05 CALCIUM CHANNELS Benzamil 0.83 0.08 0.83 0.12 0.96 0.04 CALCIUM CHANNELS BAY K-8644 0.93 0.13 0.93 0.09 1.07 0.14 CALCIUM CHANNELS Diltiazem 0.96 0.07 0.99 0.12 0.94 0.14 CALCIUM CHANNELS L-cis-Diltiazem 0.91 0.17 1.01 0.12 0.95 0.12 CALCIUM CHANNELS Flunarizine 0.85 0.08 1.00 0.06 0.85 0.05 CALCIUM CHANNELS FPL-64176 0.99 0.11 0.95 0.07 1.05 0.05 CALCIUM CHANNELS Nifedipine 1.06 0.17 0.95 0.12 1.03 0.09 CALCIUM CHANNELS Nimodipine 1.05 0.06 0.95 0.03 1.06 0.17 CALCIUM CHANNELS Nitrendipine 0.99 0.07 0.96 0.10 1.04 0.09 CALCIUM CHANNELS SDZ-202791 R(-) 1.01 0.08 0.92 0.06 1.01 0.08 CALCIUM CHANNELS SKF-96365 0.73 0.05 0.70 0.11 0.69 0.04 CALCIUM CHANNELS Tetrandrine 0.47 0.07 0.76 0.16 0.87 0.20 CALCIUM CHANNELS Verapamil 1.01 0.02 0.89 0.07 1.06 0.20 CALCIUM CHANNELS Methoxy Verapamil 0.93 0.14 0.96 0.07 0.93 0.13 CALCIUM CHANNELS Bepridil 0.70 0.16 0.92 0.15 0.84 0.14 CALCIUM CHANNELS Amiodarone 0.32 0.12 0.58 0.07 0.48 0.23 CALCIUM CHANNELS YS035 1.00 0.16 -

Impact of Itopride and Domperidone on Sensitivity of Gastric Distention and Gastric Accommodation in Healthy Volunteers
Impact of Itopride and Domperidone on sensitivity of gastric distention and gastric accommodation in healthy volunteers. Karen Van den Houte, Florencia Carbone, Ans Pauwels, Rita Vos, Tim Vanuytsel, Jan Tack Translational Research Center for Gastrointestinal Disorders, KULeuven, Belgium BACKGROUND & AIM RESULTS Functional dyspepsia (FD), defined as upper abdominal symptoms affecting Conduct of the study Gastric accommodation daily life, as postprandial fullness, early satiation, epigastric pain, and epigastric • 15 healthy volunteers (9 female, 6 male) • No significant differences in VAS scores before and after meal burning, without any underlying organic disease, is one of the most common • Mean age: 28.3±5.8 years ingestion and no significant differences in preprandial intragastric functional gastrointestinal disorders (1). volumes were observed. Itopride, a prokinetic drug with dopamine D2-antagonistic and cholinesterase Gastric compliance and gastric sensitivity • Postprandial gastric volumes and gastric accommodation were inhibitor properties, is frequently used to treat functional dyspepsia. Its effects I50, I100, and D10 did not affect fasting or postprandial gastric significantly lower for I50 and for D10, compared to placebo. on gastric sensitivity and accommodation are unknown (2), compliance and gastric sensitivity to distention significantly The aim of this study is to evaluate the effect of Itopride, compared to compared to placebo. Domperidone, a dopamine D2 receptor antagonist, on the sensitivity to gastric * Placebo balloon -

Brimonidine Tartrate; Brinzolamide
Contains Nonbinding Recommendations Draft Guidance on Brimonidine Tartrate ; Brinzolamide This draft guidance, when finalized, will represent the current thinking of the Food and Drug Administration (FDA, or the Agency) on this topic. It does not establish any rights for any person and is not binding on FDA or the public. You can use an alternative approach if it satisfies the requirements of the applicable statutes and regulations. To discuss an alternative approach, contact the Office of Generic Drugs. Active Ingredient: Brimonidine tartrate; Brinzolamide Dosage Form; Route: Suspension/drops; ophthalmic Strength: 0.2%; 1% Recommended Studies: One study Type of study: Bioequivalence (BE) study with clinical endpoint Design: Randomized (1:1), double-masked, parallel, two-arm, in vivo Strength: 0.2%; 1% Subjects: Males and females with chronic open angle glaucoma or ocular hypertension in both eyes. Additional comments: Specific recommendations are provided below. ______________________________________________________________________________ Analytes to measure (in appropriate biological fluid): Not applicable Bioequivalence based on (95% CI): Clinical endpoint Additional comments regarding the BE study with clinical endpoint: 1. The Office of Generic Drugs (OGD) recommends conducting a BE study with a clinical endpoint in the treatment of open angle glaucoma and ocular hypertension comparing the test product to the reference listed drug (RLD), each applied as one drop in both eyes three times daily at approximately 8:00 a.m., 4:00 p.m., and 10:00 p.m. for 42 days (6 weeks). 2. Inclusion criteria (the sponsor may add additional criteria): a. Male or nonpregnant females aged at least 18 years with chronic open angle glaucoma or ocular hypertension in both eyes b. -

Two Phase 3 Clinical Trials Comparing the Safety and Efficacy of Netarsudil to Timolol in Patients with Elevated Intraocular
Two Phase 3 Clinical Trials Comparing the Safety and Efficacy of Netarsudil to Timolol in Patients With Elevated Intraocular Pressure: Rho Kinase Elevated IOP Treatment Trial 1 and 2 (ROCKET-1 and ROCKET-2) JANET B. SERLE, L. JAY KATZ, EUGENE MCLAURIN, THERESA HEAH, NANCY RAMIREZ-DAVIS, DALE W. USNER, GARY D. NOVACK, AND CASEY C. KOPCZYNSKI, FOR THE ROCKET-1 AND ROCKET-2 STUDY GROUPS PURPOSE: To evaluate the efficacy and ocular and sys- ROCKET-2) for timolol (P < .0001 for netarsudil vs temic safety of netarsudil 0.02% ophthalmic solution, a timolol). rho-kinase inhibitor and norepinephrine transporter in- CONCLUSIONS: In 2 large, randomized, double-masked hibitor, in patients with open-angle glaucoma and ocular trials reported here, once-daily dosing of netarsudil hypertension. 0.02% was found to be effective and well tolerated for DESIGN: Double-masked, randomized noninferiority the treatment of patients with ocular hypertension and clinical trials: Rho Kinase Elevated IOP Treatment Trial open-angle glaucoma. The novel pharmacology and 1 and 2 (ROCKET-1 and ROCKET-2). aqueous humor dynamic effects of this molecule suggest METHODS: After a washout of all pre-study ocular hy- it may be a useful addition to the armamentarium of potensive medications, eligible patients were randomized ocular hypotensive medications. (Am J Ophthalmol to receive netarsudil 0.02% once daily (q.d.), timolol 2018;186:116–127. Ó 2017 The Authors. Published 0.5% twice a day (b.i.d.), and (ROCKET-2 only) netar- by Elsevier Inc. This is an open access article under the sudil 0.02% b.i.d. -

PDE4-Inhibitors: a Novel, Targeted Therapy for Obstructive Airways Disease Zuzana Diamant, Domenico Spina
PDE4-inhibitors: A novel, targeted therapy for obstructive airways disease Zuzana Diamant, Domenico Spina To cite this version: Zuzana Diamant, Domenico Spina. PDE4-inhibitors: A novel, targeted therapy for obstructive airways disease. Pulmonary Pharmacology & Therapeutics, 2011, 24 (4), pp.353. 10.1016/j.pupt.2010.12.011. hal-00753954 HAL Id: hal-00753954 https://hal.archives-ouvertes.fr/hal-00753954 Submitted on 20 Nov 2012 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Accepted Manuscript Title: PDE4-inhibitors: A novel, targeted therapy for obstructive airways disease Authors: Zuzana Diamant, Domenico Spina PII: S1094-5539(11)00006-X DOI: 10.1016/j.pupt.2010.12.011 Reference: YPUPT 1071 To appear in: Pulmonary Pharmacology & Therapeutics Received Date: 2 October 2010 Revised Date: 5 December 2010 Accepted Date: 24 December 2010 Please cite this article as: Diamant Z, Spina D. PDE4-inhibitors: A novel, targeted therapy for obstructive airways disease, Pulmonary Pharmacology & Therapeutics (2011), doi: 10.1016/j.pupt.2010.12.011 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. -

Properties and Units in Clinical Pharmacology and Toxicology
Pure Appl. Chem., Vol. 72, No. 3, pp. 479–552, 2000. © 2000 IUPAC INTERNATIONAL FEDERATION OF CLINICAL CHEMISTRY AND LABORATORY MEDICINE SCIENTIFIC DIVISION COMMITTEE ON NOMENCLATURE, PROPERTIES, AND UNITS (C-NPU)# and INTERNATIONAL UNION OF PURE AND APPLIED CHEMISTRY CHEMISTRY AND HUMAN HEALTH DIVISION CLINICAL CHEMISTRY SECTION COMMISSION ON NOMENCLATURE, PROPERTIES, AND UNITS (C-NPU)§ PROPERTIES AND UNITS IN THE CLINICAL LABORATORY SCIENCES PART XII. PROPERTIES AND UNITS IN CLINICAL PHARMACOLOGY AND TOXICOLOGY (Technical Report) (IFCC–IUPAC 1999) Prepared for publication by HENRIK OLESEN1, DAVID COWAN2, RAFAEL DE LA TORRE3 , IVAN BRUUNSHUUS1, MORTEN ROHDE1, and DESMOND KENNY4 1Office of Laboratory Informatics, Copenhagen University Hospital (Rigshospitalet), Copenhagen, Denmark; 2Drug Control Centre, London University, King’s College, London, UK; 3IMIM, Dr. Aiguader 80, Barcelona, Spain; 4Dept. of Clinical Biochemistry, Our Lady’s Hospital for Sick Children, Crumlin, Dublin 12, Ireland #§The combined Memberships of the Committee and the Commission (C-NPU) during the preparation of this report (1994–1996) were as follows: Chairman: H. Olesen (Denmark, 1989–1995); D. Kenny (Ireland, 1996); Members: X. Fuentes-Arderiu (Spain, 1991–1997); J. G. Hill (Canada, 1987–1997); D. Kenny (Ireland, 1994–1997); H. Olesen (Denmark, 1985–1995); P. L. Storring (UK, 1989–1995); P. Soares de Araujo (Brazil, 1994–1997); R. Dybkær (Denmark, 1996–1997); C. McDonald (USA, 1996–1997). Please forward comments to: H. Olesen, Office of Laboratory Informatics 76-6-1, Copenhagen University Hospital (Rigshospitalet), 9 Blegdamsvej, DK-2100 Copenhagen, Denmark. E-mail: [email protected] Republication or reproduction of this report or its storage and/or dissemination by electronic means is permitted without the need for formal IUPAC permission on condition that an acknowledgment, with full reference to the source, along with use of the copyright symbol ©, the name IUPAC, and the year of publication, are prominently visible. -

Table 1. Glaucoma Medications: Mechanisms, Dosing and Precautions Brand Generic Mechanism of Action Dosage/Avg
OPTOMETRIC STUDY CENTER Table 1. Glaucoma Medications: Mechanisms, Dosing and Precautions Brand Generic Mechanism of Action Dosage/Avg. % Product Sizes Side Effects Warnings Reduction CHOLINERGIC AGENTS Direct Pilocarpine (generic) Pilocarpine 1%, 2%, 4% Increases trabecular outflow BID-QID/15-25% 15ml Headache, blurred vision, myopia, retinal detachment, bronchiole constriction, Angle closure, shortness of breath, retinal narrowing of angle detachment Indirect Phospholine Iodide (Pfizer) Echothiophate iodide 0.125% Increases trabecular outflow QD-BID/15-25% 5ml Same as above plus cataractogenic iris cysts in children, pupillary block, Same as above, plus avoid prior to any increased paralysis with succinylcholine general anesthetic procedure ALPHA-2 AGONISTS Alphagan P (Allergan) Brimonidine tartrate 0.1%, 0.15% with Purite Decreases aqueous production, increases BID-TID/up to 26% 5ml, 10ml, 15ml Dry mouth, hypotension, bradycardia, follicular conjunctivitis, ocular irritation, Monitor for shortness of breath, dizziness, preservative uveoscleral outflow pruritus, dermatitis, conjunctival blanching, eyelid retraction, mydriasis, drug ocular redness and itching, fatigue allergy Brimonidine tartrate Brimonidine tartrate 0.15%, 0.2% Same as above Same as above 5ml, 10ml Same as above Same as above (generic) Iopidine (Novartis) Apraclonidine 0.5% Decreases aqueous production BID-TID/up to 25% 5ml, 10ml Same as above but higher drug allergy (40%) Same as above BETA-BLOCKERS Non-selective Betagan (Allergan) Levobunolol 0.25%, 0.5% Decreases -

Customs Tariff - Schedule
CUSTOMS TARIFF - SCHEDULE 99 - i Chapter 99 SPECIAL CLASSIFICATION PROVISIONS - COMMERCIAL Notes. 1. The provisions of this Chapter are not subject to the rule of specificity in General Interpretative Rule 3 (a). 2. Goods which may be classified under the provisions of Chapter 99, if also eligible for classification under the provisions of Chapter 98, shall be classified in Chapter 98. 3. Goods may be classified under a tariff item in this Chapter and be entitled to the Most-Favoured-Nation Tariff or a preferential tariff rate of customs duty under this Chapter that applies to those goods according to the tariff treatment applicable to their country of origin only after classification under a tariff item in Chapters 1 to 97 has been determined and the conditions of any Chapter 99 provision and any applicable regulations or orders in relation thereto have been met. 4. The words and expressions used in this Chapter have the same meaning as in Chapters 1 to 97. Issued January 1, 2020 99 - 1 CUSTOMS TARIFF - SCHEDULE Tariff Unit of MFN Applicable SS Description of Goods Item Meas. Tariff Preferential Tariffs 9901.00.00 Articles and materials for use in the manufacture or repair of the Free CCCT, LDCT, GPT, UST, following to be employed in commercial fishing or the commercial MT, MUST, CIAT, CT, harvesting of marine plants: CRT, IT, NT, SLT, PT, COLT, JT, PAT, HNT, Artificial bait; KRT, CEUT, UAT, CPTPT: Free Carapace measures; Cordage, fishing lines (including marlines), rope and twine, of a circumference not exceeding 38 mm; Devices for keeping nets open; Fish hooks; Fishing nets and netting; Jiggers; Line floats; Lobster traps; Lures; Marker buoys of any material excluding wood; Net floats; Scallop drag nets; Spat collectors and collector holders; Swivels. -

)&F1y3x PHARMACEUTICAL APPENDIX to THE
)&f1y3X PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE )&f1y3X PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 3 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. Product CAS No. Product CAS No. ABAMECTIN 65195-55-3 ACTODIGIN 36983-69-4 ABANOQUIL 90402-40-7 ADAFENOXATE 82168-26-1 ABCIXIMAB 143653-53-6 ADAMEXINE 54785-02-3 ABECARNIL 111841-85-1 ADAPALENE 106685-40-9 ABITESARTAN 137882-98-5 ADAPROLOL 101479-70-3 ABLUKAST 96566-25-5 ADATANSERIN 127266-56-2 ABUNIDAZOLE 91017-58-2 ADEFOVIR 106941-25-7 ACADESINE 2627-69-2 ADELMIDROL 1675-66-7 ACAMPROSATE 77337-76-9 ADEMETIONINE 17176-17-9 ACAPRAZINE 55485-20-6 ADENOSINE PHOSPHATE 61-19-8 ACARBOSE 56180-94-0 ADIBENDAN 100510-33-6 ACEBROCHOL 514-50-1 ADICILLIN 525-94-0 ACEBURIC ACID 26976-72-7 ADIMOLOL 78459-19-5 ACEBUTOLOL 37517-30-9 ADINAZOLAM 37115-32-5 ACECAINIDE 32795-44-1 ADIPHENINE 64-95-9 ACECARBROMAL 77-66-7 ADIPIODONE 606-17-7 ACECLIDINE 827-61-2 ADITEREN 56066-19-4 ACECLOFENAC 89796-99-6 ADITOPRIM 56066-63-8 ACEDAPSONE 77-46-3 ADOSOPINE 88124-26-9 ACEDIASULFONE SODIUM 127-60-6 ADOZELESIN 110314-48-2 ACEDOBEN 556-08-1 ADRAFINIL 63547-13-7 ACEFLURANOL 80595-73-9 ADRENALONE