Comparison of Glaucoma-Relevant Transcriptomic Datasets Identifies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

COUNTRY BASED ADMISSION CRITERIA Country Examination Minimum Criteria INTERNATIONAL SAT (Scholastic Assessment Test) Reasoning
COUNTRY BASED ADMISSION CRITERIA Country Examination Minimum criteria INTERNATIONAL SAT (Scholastic Assessment Test) Min. 1000 Reasoning. KADIR HAS INSTITUTIONAL CODE: 7396 INTERNATIONAL Abitur and Fachabitür Max.4 INTERNATIONAL Al-Shahada-Al Thanawiyya For all B.Sc. Min. 180, for B.A. programs min. 170 INTERNATIONAL Al-Shahada-Al Thanawiyya Al- For all B.Sc. Min. 180, for Amma B.A. programs min. 170 INTERNATIONAL American College Test (ACT) Math, Science and total Min. 21 INTERNATIONAL Attestat o Srednem Min. 3 (Polinom) Obshchem Obrazovanii INTERNATIONAL French Baccalaureate Min. 12 INTERNATIONAL GCE (General Certificate of Min. 2 A-levels with grades D and Education) above INTERNATIONAL IB (International Baccalaureate) min. 28 INTERNATIONAL Matura Max.4 INTERNATIONAL Tawjihi For all B.Sc. Min. 80, for B.A. programs min. 75 INTERNATIONAL International Student Science Olympics Golden, Silver, or Bronze Medals from the international student Olympics recognized by TUBITAK INTERNATIONAL West African Examination Council- Min. C grade from min. 6 subjects International Senior School Certificate Examination (WEAC-SSCE) INTERNATIONAL YÖS Min. 50 INTERNATIONAL European Baccalaureate Min. 60% INTERNATIONAL BTEC – QSF Diploma Min. D grades from min. 3 subjects INTERNATIONAL Western European Open School Min. 60% Examination INTERNATIONAL International High School Diploma Min. 60% AUSTRIA Maturazeugnis/Matura/Reifeprufungsze Min. 3 ugnis /Abschlusszeugnis from a Fachschule (with at least 4 years of full time study) AFGHANISTAN KONKUR General State Examination Min. 210/350 or 60% or High School Diploma (Doreyeh Aali) ALBANIA Dëftesë Pjekurie / Certificate of Min. 6 Maturity ALGERIA Algerian baccalaureate Min. 12 ANGOLA Diploma de Ensino Pre Universitario / Min. 12 Carta do Curso Complementar Liceus/Habilitacoes Literarias ARGENTINA Titulo de Bachiller Min. -

International-Vwo-Equivalent-Diplomas-2014-2015.Pdf
Minimum entrance qualifications for non-Dutch diplomas Maastricht University 2014/2015 Country Diploma International European Baccalaureate Diploma schools International Baccalaureate Diploma Be aware! A Certificate is not sufficient! Albania Dëftese Pjekurie Austria Reifezeugnis / Reifeprüfungszeugnis Azerbaijan Orta Tahsil Haqqinda Attestat (average grade of 4) and national entrance examination Belgium Diploma van Secundair Onderwijs / Diplome de l'Énseignement Secondaire Superieur / Abschlusszeugnis der Oberstufe des Sekundarunterrichts Brazil Certificado de Conclusao de 2° grau and 1 completed year university education Bosnia- Matura Herzegovina Bulgaria Diploma za Zavarsheno Sredno Obrazovanie (Diploma za Zavarsheno Sredno Obrazovanie) Canada, Ontario OSSD including at least 6 U-courses or OAC’s Canada, Quebec Diploma d’Etudes Collégiales (DEC) préuniversitaire Canada, other High School Diploma with a relatively large number of academic courses in grades 11 and 12 with good grades China Senior Middle School Graduation Certificate (Gaozhong) and 1 completed year university education Croatia Svjedodžba o (državnoj) maturi obtained at a Gymnazijum Cyprus Apolytirio - Gymnasium Czech republic Vysvedcení o Maturitní Zkoušce obtained at a Gymnázium Denmark Studentereksamenbevis (STX) or Brevis for Høgere Forberedelseksamen (HF) Estonia Gümnaasiumi Loputunnistus Finland Ylioppilastutkintotodistus France Diplome du Baccalauréat Général or Diplôme du Baccalauréat Technologique. Germany Zeugnis der Allgemeinen Hochschulreife (Abitur) Greece -

Matura Examinations in Slovenia Case Study of the Introduction of an External Examinations System for Schools
Matura Examinations in Slovenia Case Study of the Introduction of an External Examinations System for Schools – Sergij Gabrøœek – George Bethell CIP – Kataloæni zapis o publikaciji Narodna in univerzitetna knjiænica, Ljubljana 371.274/.276:373.5 GABRØŒEK, Sergij Matura examinations in Slovenia : case study of the introduction of an external examinations system for schools / Sergij Gabrøœek, George Bethell. – Ljubljana : National Examinations Centre, 1996 ISBN 961-6120-49-2 1. Bethell, George 61917696 Matura Examinations in Slovenia Case Study of the Introduction of an External Examinations System for Schools – Sergij Gabrøœek1 – George Bethell 2 1 Dr. Sergij Gabrøœek, Director, National Examinations Centre, Podmiløœakova 25, 1000 Ljubljana, Slovenia 2 George Bethell, Educational Consultant, 17 Orchard Avenue, Cambridge CB4 2AQ, United Kingdom Contents 7 – Abstract 9 – Background to Slovenia 11 – History of the Development of Matura 21 Description of Matura and Comparison with the School-Based – ‘Final Examination’ 25 – Developing Subject Catalogues for Matura 29 – Development of Question Papers 33 – Preparing for Matura: Trial Examinations 41 – Preparing for Matura: Gaining Support 45 – The National Examinations Centre 53 – Future Activities 57 – Conclusions and Analysis 7 Abstract The place of assessment and certification of learner achievement continues to be of great interest throughout the world. However, in eastern and central Europe there is a particular urgency about the debate as newly independent states review and reform their education systems in the light of changing social and economic conditions. Slovenia is currently in the middle of a development programme which will eventually affect all parts of its education system. In July 1995, a major feature of the programme became reality when the first Matura, or graduation examination, was successfully conducted on a national scale. -
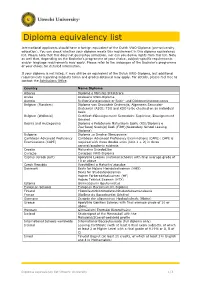
Diploma Equivalency List
Diploma equivalency list International applicants should have a foreign equivalent of the Dutch VWO-Diploma (pre-university education). You can check whether your diploma meets this requirement in this diploma equivalency list. Please note that this does not guarantee admission, nor can you derive rights from this list. Note as well that, depending on the Bachelor’s programme of your choice, subject-specific requirements and/or language requirements may apply. Please refer to the webpages of the Bachelor's programme of your choice for detailed information. If your diploma is not listed, it may still be an equivalent of the Dutch VWO-Diploma, but additional requirements regarding subjects taken and grades obtained may apply. For details, please feel free to contact the Admissions Office. Country Name Diploma Albania Diplomë e Maturës Shtetërore Aruba Arubaans VWO-Diploma Austria Reifeprüfungszeugnis or Reife- und Diplomprüfungszeugnis Belgium (Flanders) Diploma van Secundair Onderwijs, Algemeen Secundair Onderwijs (ASO); TSO and KSO to be checked on an individual basis Belgium (Wallonia) Certificat d'Enseignement Secondaire Supérieur, Enseignement Général Bosnia and Herzegovina Diploma o Položenom Maturskom Ispitu (RS)/Diploma o Završenoj Srednjoj školi (FiBH)(Secondary School Leaving Diploma) Bulgaria Diploma za Sredno Obrazovanie Caribbean Advanced Proficiency Caribbean Advanced Proficiency Examinations (CAPE): CAPE is Examinations (CAPE) required with three double units (Unit 1 + 2) in three general/academic subjects Croatia Maturalna -

Higher Education Entrance Qualifications and Exams in Europe: a Comparison
DIRECTORATE-GENERAL FOR INTERNAL POLICIES POLICY DEPARTMENT B: STRUCTURAL AND COHESION POLICIES CULTURE AND EDUCATION HIGHER EDUCATION ENTRANCE QUALIFICATIONS AND EXAMS IN EUROPE: A COMPARISON STUDY This document was requested by the European Parliament's Committee on Culture and Education. AUTHORS Cecile Hoareau McGrath, Marie Louise Henham, Anne Corbett, Niccolo Durazzi, Michael Frearson, Barbara Janta, Bregtje W. Kamphuis, Eriko Katashiro, Nina Brankovic, Benoit Guerin, Catriona Manville, Inga Schwartz, Daniel Schweppenstedde RESPONSIBLE ADMINISTRATOR Markus J. Prutsch Policy Department B: Structural and Cohesion Policies European Parliament B-1047 Brussels E-mail: [email protected] EDITORIAL ASSISTANCE Lyna Pärt LINGUISTIC VERSIONS Original: EN Translation: DE, FR ABOUT THE PUBLISHER To contact the Policy Department or to subscribe to its monthly newsletter please write to: [email protected] Manuscript completed in May 2014 Brussels © European Union, 2014 This document is available on the Internet at: http://www.europarl.europa.eu/studies DISCLAIMER The opinions expressed in this document are the sole responsibility of the authors and do not necessarily represent the official position of the European Parliament. Reproduction and translation for non-commercial purposes are authorized, provided the source is acknowledged and the publisher is given prior notice and sent a copy. DIRECTORATE-GENERAL FOR INTERNAL POLICIES POLICY DEPARTMENT B: STRUCTURAL AND COHESION POLICIES CULTURE AND EDUCATION HIGHER EDUCATION ENTRANCE QUALIFICATIONS AND EXAMS IN EUROPE: A COMPARISON STUDY Abstract The study analyses admission systems to higher education across ten countries, covering some countries of the European Union (France, Germany, Italy, Slovenia, Sweden and the United Kingdom), a candidate country (Turkey) as well as commonly used international comparators (Australia, Japan and the US). -

Exam Accepted Exam Details ACT ACT (American College Test) Exam
VALID EXAM AND DIPLOMA FOR APPLICATION A certificate with the status of the diploma and Matura are valid indefinitely as stated below list, Exams that have the status of the University Entrance exam are valid for the two years. 100% of the points are counted for the exams stated in Table 1 and 2. Table 1 Exam Accepted Exam Details For the Faculty of Medicine and Dentistry at least to have in total (Science), (Mathematics) 27 points out of 36 ACT ACT (American College Test) Exam For the Engineering-Architecture programs minimum 25 points, For the other undergraduate programs minimum 23 points For Associate Degree programs minimum 20 points For the Faculty of Medicine and Dentistry “Evidence-Based Reading and Writing” and “Math” for each test to have at least 640 points, SAT 1 SAT 1 For the Engineering-Architecture programs' minimum 560 points, For the other undergraduate programs minimum 480 points For Associate Degree programs minimum of 320 points GCE GCE (A LEVEL EXAM) At least to have A-Level in 3 subject which is related to the at least one (A LEVEL (Application with expected results applied program. EXAM) cannot be accepted.) Table 2 Exam Accepted Exam Details For the Faculty of Medicine and Dentistry minimum of 80 points, SAKARYA Sakarya University International For the Engineering-Architecture programs minimum 70 points, YOS Student Exam (SAKARYA YOS) For the other undergraduate programs minimum 60 points, For Associate Degree programs minimum 40 points 80% of the points are counted for the exams stated in Table 3 Table 3 Country Accepted Exam Details The other Universities YOS exam results accepted by Sakarya The exams are to be held by other higher University will be multiplied by 0.8 in placement procedure. -

ENIC NARIC Networks Compilation on the Situation of Upper Secondary
ENIC NARIC Networks Compilation on the situation of upper secondary school leaving qualifications due to the Covid-19 - March 2021 Updated on: Country Will this year’s upper secondary graduates receive a Will the final diploma have either a calculated Are there any consequences for admission to higher education in your country for this Other Comments final diploma? If yes, when? grade point average or calculated grades in each year’s graduates? subject? 18/03/21 Armenia Yes, in June Yes, it will have both. It will need special organisation of the admission process depending on the pandemic situation. 11/04/21 Cambridge Our priority is delivering exams safely and fairly, allowing Where exams cannot take place, we are In setting exam standards, we take very seriously our responsibility to make sure We continue to work with universities, so they understand our approach International learners to progress. switching from exams to school-assessed grades students are assessed fairly and can progress with their education. Exams offer for 2021 and can make informed admission and credit decisions. GCSE og GCE AS- We plan for exams to go ahead in June 2021 where it is using student work, so that students can consistent, reliable and fair measurement of attainment, and we have highly Universities will evaluate applications fairly from all students whether and A-levels (UK) permitted and safe. We know this is the fairest and most progress despite the impact of the pandemic. effective processes for managing standards in the grades from exams. We will their work is assessed by exams or by teacher assessment. -

2 Matura Exam Practice © Oxford University Press Speaking Discussing a Topic 10 Topic Shopping and Services
HeadwayNew Matura Exam Practice and Culture & Literature Companion Basic Level 2 HeadwayNew Matura Exam Practice and Culture & Literature Companion Basic Level 3 Contents Matura Exam Practice 1 Reading Multiple matching People p4 2 Listening Multiple matching Education p5 3 Writing Informal letter Hospitality and food p6 4 Speaking Comparing and discussing Employment p7 5 Use of English Word formation Culture and sport p8 6 Reading Matching People p9 7 Reading Open cloze Shopping and services p10 8 Listening Multiple choice Science and technology p11 9 Reading Multiple choice State and society p12 10 Speaking Discussing a topic Shopping and services p13 11 Writing Discursive essays State and society p14 12 Reading Matching Travel and tourism p15 13 Use of English Multiple choice cloze Culture p16 14 Reading Multiple choice Travel and tourism p17 15 Writing Notes and messages People p18 16 Reading Gap fill Culture p19 17 Listening Multiple choice Leisure p20 18 Listening Multiple choice Children p21 Culture & Literature 1 Culture The British Empire p22 2 Culture The Globe Theatre p24 3 Culture Education in the UK and the US p26 4 Culture Super size America; super size world? p28 5 Culture English-speaking capitals p30 6 Culture Australia: Going to live Down Under p32 7 Culture Transport in London p34 8 Literature Sir Arthur Conan Doyle – The Hound of the Baskervilles p36 Matura Exam Practice Answer Key p38 Matura Exam Practice Tapescripts p41 Culture & Literature Answer Key p43 Culture & Literature Glossary p46 ReaDING Multiple matching 1 TOPIC People 3 In pairs, write a word or phrase to summarize the EXAM TIPS meaning of the sentences (1–5) below. -

International Student Admissions Requirements
International Admissions Requirements If you’re studying outside of Canada, please use this document to determine the relevant country requirements for eligibility. Common international curriculums are listed on page one. subjects OR three different Common International Advanced Level (A Baccalaureate (IB) Level)/International International Advanced Level academic Minimum Admission subjects. Curriculums Requirements: The International Baccalaureate diploma including English HL French-Patterned U.S.-Patterned or SL. Most successful Education Education applicants present a predicted score of 27 or Minimum Admission Minimum Admission higher. IB Certificates may be Requirements: Baccalauréat Requirements: Senior considered in certain /Baccalauréat Year/Grade 12 in an circumstances. Genéral/Diplôme de accredited high school with a Bachelier de l’Enseignement calculated grade point Transfer credit may be du Second Degré/Option average and sufficient awarded for your Higher Internationale du breadth of courses. Level subjects. Baccalauréat. Most successful applicants Caribbean Advanced West African present a grade point Proficiency Examinations Council average of C+ or higher. Examination (CAPE) (WAEC) Transfer credit may be awarded for your Advanced Minimum Admission Minimum Admission Placement (AP) Requirements: The Requirements: complete diploma (six units) examinations. West African Examinations offered by the Caribbean Examinations Council. Council Senior School U.S. students completing Certificate Examination alternative curriculum are -

Covid-19 Info on Upper Secondary School Exams
Draft ENIC NARIC Network Compilation of data on upper secondary arrangements prepared by the network June-July 2020 Date updated Country Question Question Question 3 Question 4 Will this year’s upper Will the final Diploma have a Are there any consequences for Other Comments secondary graduates receive a calculated point average? admission to higher education in final diploma? If yes, when? your country? 29/06/20 Albania As per Albanian Ministry of Education As per info given by our Ministry of No, there will be no consequences for Taking into consideration the decision the Maturity exams will take Education the Diploma will have a admission to higher education. optimistic situation about Covid 19 place during the June 2020. Maturity calculated grade average. As the grade in Albania, and the facts that the Diploma will be issued within July average is stipulated at Decision of restrictions are eased since 27 th of 2020. Council of Ministers, which for this April 2020, the government has year is still in the process of approval, decided to open the schools in we will confirm as soon as approved. Albania for maturity classes by 18th of May 2020. 10/07/20 Andorra Upper secondary students will receive a Yes, students are having a calculated There are no special consequences for final diploma issued at the end of June grade point average. Two different admission to higher education in 2020. decisions are in place when it comes Andorra for this year's graduates. to calculate the grade point average: 1. Students graduating from general upper secondary program are having a grade calculated in the last tests based on competences. -

Guidelines Mathematics Requirements 2021-2022 Liberal Arts & Sciences
Leiden University College The Hague Admissions Office [email protected] Guidelines mathematics requirements 2021-2022 Liberal Arts & Sciences: Global Challenges All students interested in the LUC programme must have mathematics skills in order to pass the compulsory mathematics course in the first year. Check the information below to determine if you have mathematics at the minimum level required for admission. Please note: The fact that a diploma and/or Mathematics level/grade is stated does not mean your mathematics skills are sufficient. This list provides minimum requirements and the Admissions Committee can always determine that an additional LUC mathematics course is required. Applicants who meet the mathematics requirement or still need to graduate and are likely to meet the Mathematics requirement, will have a higher chance to be selected for the programme. Please click on the diploma name or country to go to the specific mathematics requirements. (International) Diplomas VWO Diploma - Netherlands Cambridge or Pearson International Examinations Caribbean Advanced Proficiency Examination (CAPE) European Baccalaureate International Baccalaureate (IB) Boswell-Bèta - certificate US high school diploma from an international school outside the US Foreign/national diplomas Algeria Denmark Japan Poland Turkey Argentina Ecuador Jordan Portugal Ukraine Aruba Estonia Korea, South Romania United Kingdom Australia Finland Latvia Russian Federation United States Austria France Lithuania Serbia Venezuela Belgium Georgia Luxembourg -

Country Qualifications Grade Range Equivalence ` Subject Equivalence - A'level
COUNTRY QUALIFICATIONS GRADE RANGE EQUIVALENCE ` SUBJECT EQUIVALENCE - A'LEVEL A-levels A*A*A* A*A*A A*AA AAA AAB ABB BBB BBC A* A B C Austria Reifeprufungszeugnis/Maturazeugnis Pass your Pass your Pass your Pass your Pass your Pass your Pass your Pass your 1 1 2 Reifeprufung mit Reifeprufung mit Reifeprufung mit Reifeprufung mit Reifeprufung mit Reifeprufung mit Reifeprufung Reifeprufung ausgezeichnetem ausgezeichnetem ausgezeichnete ausgezeichnete gutem Erfolg gutem Erfolg mit gutem Bestanden, with Erfolg, with four Erfolg bestanden, m Erfolg, m Erfolg bestanden, with bestanden, with Erfolg threer subjects subjects(written with four bestanden with bestanden, with three one bestanden , with (written exam) at exams) at 1/sehr gut subjects(written four three subjects(written subject(written one 2/gut and one exams) at 1/sehr subjects(written subjects(written exams) at exams) at subject(written subject (wriiten gut exams) at exams) at 1/sehr gut and 1/sehr gut and exams) at exam) at 1/sehr gut 1/sehr gut and one subject three subjects 1/sehr gut and 3/Befriedigend one subject (written exam) (written exam) three (written exam) at 2/gut at 2/gut subjects(written at 2/gut exam) at 2/gut Belgium Certificat d'Enseignement Secondaire 10/10 or 20/20 or 9.5/10 or 18.5/20 9/10 or 18/20 or 8.5/10 or 8/10 or 16/20 or 7.5/10 or 7/10 or 14/20 or 6.5/10 or 9/10 or 8/10 or 7/10 or 14/20 Supérieur/ Diploma van Secundair 90% overall or 85% overall 80% overall 16.5/20 or 78% 75% overall 14.5/20 or 73% 70% overall 13.5/20 or 68% 18/20 16/20 Onderwijs overall