DEA Annual Report 2011
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

History of Badminton
Facts and Records History of Badminton In 1873, the Duke of Beaufort held a lawn party at his country house in the village of Badminton, Gloucestershire. A game of Poona was played on that day and became popular among British society’s elite. The new party sport became known as “the Badminton game”. In 1877, the Bath Badminton Club was formed and developed the first official set of rules. The Badminton Association was formed at a meeting in Southsea on 13th September 1893. It was the first National Association in the world and framed the rules for the Association and for the game. The popularity of the sport increased rapidly with 300 clubs being introduced by the 1920’s. Rising to 9,000 shortly after World War Π. The International Badminton Federation (IBF) was formed in 1934 with nine founding members: England, Ireland, Scotland, Wales, Denmark, Holland, Canada, New Zealand and France and as a consequence the Badminton Association became the Badminton Association of England. From nine founding members, the IBF, now called the Badminton World Federation (BWF), has over 160 member countries. The future of Badminton looks bright. Badminton was officially granted Olympic status in the 1992 Barcelona Games. Indonesia was the dominant force in that first Olympic tournament, winning two golds, a silver and a bronze; the country’s first Olympic medals in its history. More than 1.1 billion people watched the 1992 Olympic Badminton competition on television. Eight years later, and more than a century after introducing Badminton to the world, Britain claimed their first medal in the Olympics when Simon Archer and Jo Goode achieved Mixed Doubles Bronze in Sydney. -

Women's Doubles Results Gold Silver Bronze Bronze World Championships Yang Wei / Zhang Jiewen Gao Ling / Huang Sui Wei Yili / Zhang Yawen Kumiko Ogura / Reiko Shiota
⇧ 2008 Back to Badzine Results Page 2007 Women's Doubles Results Gold Silver Bronze Bronze World Championships Yang Wei / Zhang Jiewen Gao Ling / Huang Sui Wei Yili / Zhang Yawen Kumiko Ogura / Reiko Shiota Super Series Malaysia Open Gao Ling / Huang Sui Vita Marissa / Gresya Polii Hwang Yu Mi / Kim Min Jung Kumiko Ogura / Reiko Shiota Korea Open Gao Ling / Huang Sui Yang Wei / Zhang Jiewen Lee Hyo Jung / Lee Kyung Won Wei Yili / Zhang Yawen All England Wei Yili / Zhang Yawen Yang Wei / Zhang Jiewen Gao Ling / Huang Sui Chin Eei Hui / Wong Pei Tty Swiss Open Yang Wei / Zhao Tingting Lee Hyo Jung / Lee Kyung Won Cheng Wen Hsing / Chien Yu Chin Vita Marissa / Gresya Polii Singapore Open Wei Yili / Zhang Yawen Yang Wei / Zhao Tingting Lee Hyo Jung / Lee Kyung Won Gao Ling / Zhang Jiewen Indonesia Open Du Jing / Yu Yang Yang Wei / Zhao Tingting Gao Ling / Zhang Jiewen Gail Emms / Donna Kellogg China Masters Vita Marissa / Liliyana Natsir Yang Wei / Zhao Tingting Du Jing / Yu Yang Gail Emms / Donna Kellogg Japan Open Yang Wei / Zhang Jiewen Yu Yang / Zhao Tingting Wei Yili / Zhang Yawen Gao Ling / Huang Sui Denmark Open Yang Wei / Zhang Jiewen Lee Hyo Jung / Lee Kyung Won Yu Yang / Zhao Tingting Gail Emms / Donna Kellogg French Open Yang Wei / Zhang Jiewen Yu Yang / Zhao Tingting Cheng Wen Hsing / Chien Yu Chin Jo Novita / Gresya Polii China Open Gao Ling / Zhao Tingting Du Jing / Yu Yang Wei Yili / Zhang Yawen Pan Pan / Tian Qing Hong Kong Open Du Jing / Yu Yang Wei Yili / Zhang Yawen Cheng Wen Hsing / Chien Yu Chin Gao Ling / Zhao Tingting -
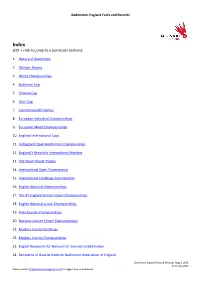
Facts and Records
Badminton England Facts and Records Index (cltr + click to jump to a particular section): 1. History of Badminton 2. Olympic Games 3. World Championships 4. Sudirman Cup 5. Thomas Cup 6. Uber Cup 7. Commonwealth Games 8. European Individual Championships 9. European Mixed Championships 10. England International Caps 11. All England Open Badminton Championships 12. England’s Record in International Matches 13. The Stuart Wyatt Trophy 14. International Open Tournaments 15. International Challenge Tournaments 16. English National Championships 17. The All England Seniors’ Open Championships 18. English National Junior Championships 19. Inter-County Championships 20. National Leisure Centre Championships 21. Masters County Challenge 22. Masters County Championships 23. English Recipients for Honours for Services to Badminton 24. Recipients of Awards made by Badminton Association of England Badminton England Facts & Records: Page 1 of 86 As at May 2021 Please contact [email protected] to suggest any amendments. Badminton England Facts and Records 25. English recipients of Awards made by the Badminton World Federation 1. The History of Badminton: Badminton House and Estate lies in the heart of the Gloucestershire countryside and is the private home of the 12th Duke and Duchess of Beaufort and the Somerset family. The House is not normally open to the general public, it dates from the 17th century and is set in a beautiful deer park which hosts the world-famous Badminton Horse Trials. The Great Hall at Badminton House is famous for an incident on a rainy day in 1863 when the game of badminton was said to have been invented by friends of the 8th Duke of Beaufort. -

2010 Training Sessions April 29-30, 2010 2010 Annual Meeting
National Council on Measurement in Education 2010 Training Sessions April 2 9-30, 2 010 2010 Annual Meeting May 1-3, 2 010 Denver, Colorado 1 NCME • 2010 Annual Meeting & Training Sessions NCME Officers President Terry A. Ackerman University of North Carolina at Greensboro President-Elect Wayne Camara The College Board Past President Mark Reckase Michigan State University Executive Officer Plumer Lovelace NCME Executive Director NCME Directors Todd Rogers University of Alberta Larry Rudner Graduate Management Admission Council Kadriye Ercikan University of British Columbia Susan Loomis National Assessment Governing Board Michael Rodriguez University of Minnesota Sherry Rose-Bond Columbus City Schools 2 Denver, Colorado Editors Journal of Educational Measurement James Carlson Educational Testing Service Educational Measurement Jacqueline Leighton Issues and Practice University of Alberta NCME Newsletter Thanos Patelis The College Board Website Management Committee Michael Bunch Measurement Incorporated 2010 Annual Meeting Chairs Annual Meeting Program Chairs John Willse University of North Carolina at Greensboro Robert Henson University of North Carolina at Greensboro Training and Development Committee Luz Bay Chair Measured Progress, Inc. Fitness Run/Walk Directors Brian French Washington State University Jill van den Heuvel CTB/McGraw-Hill 3 NCME • 2010 Annual Meeting & Training Sessions Proposal Reviewers Alagoz, Cigdem de la Torre, Jimmy Hou, Xiaodong Lee, Guemin Albano, Anthony Demars, Christine Hu, Huiqin (Ann) Lee, Won-Chan Alexeev, Natalia Deng, Nina Huang, Xiaomin Lee, Yi-Hsuan Ali, Usama Deng, Weiling Huebner, Alan Lee, Yong-Sang Allalouf, Avi Diao, Qi Huff, Kristen Lee, Yoonsun Almusawi, Numan Dolan, Robert Huggins, Anne Leenen, Iwin Alonzo, Julie Douglas, Jeffrey Corinne Levy, Roy Arce-Ferrer, Alvaro Du, Yi Hunter, C. -

歷屆成績 Champions of the Hong Kong Opens
香港公開賽歷屆成績 CHAMPIONS OF THE HONG KONG OPENS 年份 獎金 男子單打 女子單打 男子雙打 女子雙打 混合雙打 Year Prize Men’s Singles Women’s Singles Men’s Doubles Women’s Doubles Mixed Doubles 1982 HK$170,000 百加珠 (印度) 徐蓉 (中國) 陳金德/楊榮美 (印尼) 羅娜比莉/珍威絲達 (英國) Prakash (IND) Xu Rong (CHN) Karrono/Heryanto (INA) Nora Perry/J.Webster (ENG) 1985 HK$170,000 楊陽 (中國) 韓愛萍 (中國) 凱文迪/費力保 (丹麥) 徐蓉/韓愛萍 (中國) 杜絲/吉爾絲 (英國) Yang Yang (CHN) Han Aiping (CHN) Heryanto/F. Fladberg (DEN) Xu Rong/Han Aiping (CHN) Martin Dew/G. Gilks (ENG) 1986 HK$200,000 楊陽 (中國) 李玲蔚 (中國) 葉忠明/楊榮美 (印尼) 李玲蔚/韓愛萍 (中國) 格利蘭/羅娜比莉 (英國) Yang Yang (CHN) Li Lingwei (CHN) Ertano/Heryanton (INA) Li Lingwei/Han Aiping (CHN) Billy Gilliland/Nora Perry (ENG) 1987 HK$273,000 熊國寶 (中國) 韓愛萍 (中國) 張強/周金燦 (中國) 金鍊子/鄭素英 (南韓) 格利蘭/高華絲 (英國) Xiong Guobao (CHN) Han Aiping (CHN) Zhang Qiang/Zhou Jincan (CHN) Kim Yun Ja/Chung So Young (KOR) Billy Gilliland/Gillan Gowers (ENG) 1988 HK$273,000 蘇基亞圖 (印尼) 李英淑 (南韓) 李相福/李光珍 (南韓) 林瑛/關渭貞 (中國) 朴柱奉/鄭明熙 (南韓) Icuk Sugiarto (INA) Li Yong Suk (KOR) Lee Sang Bok/Lee Kwang Jin (KOR) Li Ying/Guan Weizhen (CHN) Park Joo Bong/Chung Myung Hee (KOR) 1989 HK$466,000 吳文凱 (中國) 韓愛萍 (中國) 拉昔夫/賈蘭尼西廸 (馬來西亞) 林瑛/關渭貞 (中國) 崔相範/鄭素英 (南韓) Wu Weikai (CHN) Han Aiping (CHN) Razif Sidek/Jalani Sidek (MAS) Li Ying/Guan Weizhen (CHN) Choi Sang Bun/Chung So Young (KOR) 1991 HK$466,000 劉軍 (中國) 黃華 (中國) 李相福/孫振煥 (南韓) 黃惠英/吉永雅 (南韓) 李相福/鄭素英 (南韓) Liu Jun (CHN) Huang Hua (CHN) Lee Sang Bok/Shon Jin Hwan (KOR) Hwang Hye Young/Gil Young Ah (KOR) Lee Sang Bok/Chung So Young (KOR) 1992 HK$466,000 吳文凱 (中國) 方銖賢 (南韓) 力奇/歷斯 (印尼) 農羣華/周蕾 (中國) 李相福/吉永雅 (南韓) Wu -

Olimpijske Igre (1992-2008)
OLIMPIJSKE IGRE (1992-2008) Barcelona, 1992 Men’s Singles 1. Allan Budi Kusuma (INA) 2. Ardy Bernardus Wiranata (INA) 3. Thomas Stuer-Lauridsen (DEN) Hermawan Susanto (INA) Ladies’ Singles 1. Susi Susanti (INA) 2. Bang Soo-Hyun (KOR) 3. Huang Hua (CHN) Tang Jiuhong (CHN) Men’s Doubles 1. Park Joo-Bong / Kim Moon-Soo (KOR) 2. Eddy Hartono / Rudy Gunawan (INA) 3. Rashid Sidek / Razif Sidek (MAS) Li Yongbo / Tian Bingyi (CHN) Ladies’ Doubles 1. Hwang Hae-Young / Chung So-Young (KOR) 2. Guan Weizhen / Nong Qunhua (CHN) 3. Gil Young-Ah / Shim Eun-Jung (KOR) Lin Yanfen / Yao Fen (CHN) Atlanta, 1996 Men’s Singles 1. Poul Erik Hoyer-Larsen (DEN) 2. Dong Jiong (CHN) 3. Rashid Sidek (MAS) Ladies’ Singles 1. Bang Soo-Hyun (KOR) 2. Mia Audina (INA) 3. Susi Susanti (INA) Men’s Doubles 1. Ricky Achmad Subagja / Rexy Ronald Mainaky (INA) 2. Cheah Soon Kit / Yap Kim Hock (MAS) 3. Denny Kantono / Irianto Antonius (INA) Ladies’ Doubles 1. Ge Fei / Gu Jun (CHN) 2. Gil Young-Ah / Jang Hye Ock (KOR) 3. Qin Yiyuan / Tang Yongshu (CHN) Mixed Doubles 1. Zhang Jun / Gao Ling (CHN) 2. Park Joo-Bong / Ra Kyung-Min (KOR) 3. Liu Jianjun / Sun Man (CHN) Sydney, 2000 Men’s Singles 1. Ji Xinpeng (CHN) 2. Hendrawan (INA) 3. Xia Xuanze (CHN) Ladies’ Singles 1. Gong Zhichao (CHN) 2. Camilla Martin (DEN) 3. Ye Zhaoying (CHN) Men’s Doubles 1. Tony Gunawan / Candra Wijaya (INA) 2. Lee Dong Soo / Yoo Yong-Sung (KOR) 3. Ha Tae-Kwon / Kim Dong Moon (KOR) Ladies’ Doubles 1. -
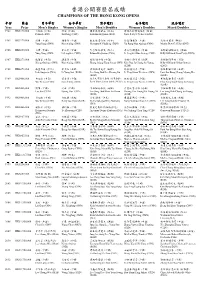
Past Result 2014
香港公開賽歷屆成績 CHAMPIONS OF THE HONG KONG OPENS 年份 獎金 男子單打 女子單打 男子雙打 女子雙打 混合雙打 Year Prize Men’s Singles Women’s Singles Men’s Doubles Women’s Doubles Mixed Doubles 1982 HK$170,000 百加珠 (印度) 徐蓉 (中國) 陳金德/楊榮美 (印尼) 羅娜比莉/珍威絲達 (英國) Prakash (IND) Xu Rong (CHN) Karrono/Heryanto (INA) Nora Perry/J.Webster (ENG) 1985 HK$170,000 楊陽 (中國) 韓愛萍 (中國) 凱文迪/費力保 (丹麥) 徐蓉/韓愛萍 (中國) 杜絲/吉爾絲 (英國) Yang Yang (CHN) Han Aiping (CHN) Heryanto/F. Fladberg (DEN) Xu Rong/Han Aiping (CHN) Martin Dew/G. Gilks (ENG) 1986 HK$200,000 楊陽 (中國) 李玲蔚 (中國) 葉忠明/楊榮美 (印尼) 李玲蔚/韓愛萍 (中國) 格利蘭/羅娜比莉 (英國) Yang Yang (CHN) Li Lingwei (CHN) Ertano/Heryanton (INA) Li Lingwei/Han Aiping (CHN) Billy Gilliland/Nora Perry (ENG) 1987 HK$273,000 熊國寶 (中國) 韓愛萍 (中國) 張強/周金燦 (中國) 金鍊子/鄭素英 (南韓) 格利蘭/高華絲 (英國) Xiong Guobao (CHN) Han Aiping (CHN) Zhang Qiang/Zhou Jincan CHN) Kim Yun Ja/Chung So Young Billy Gilliland/Gillan Gowers (KOR) (ENG) 1988 HK$273,000 蘇基亞圖 (印尼) 李英淑 (南韓) 李相福/李光珍 (南韓) 林瑛/關渭貞 (中國) 朴柱奉/鄭明熙 (南韓) Icuk Sugiarto (INA) Li Yong Suk (KOR) Lee Sang Bok/Lee Kwang Jin Li Ying/Guan Weizhen (CHN) Park Joo Bong/Chung Myung Hee (KOR) (KOR) 1989 HK$466,000 吳文凱 (中國) 韓愛萍 (中國) 拉昔夫/賈蘭尼西廸 (馬來西亞) 林瑛/關渭貞 (中國) 崔相範/鄭素英 (南韓) Wu Weikai (CHN) Han Aiping (CHN) Razif Sidek/Jalani Sidek (MAS) Li Ying/Guan Weizhen (CHN) Choi Sang Bun/Chung So Young (KOR) 1991 HK$466,000 劉軍 (中國) 黃華 (中國) 李相福/孫振煥 (南韓) 黃惠英/吉永雅 (南韓) 李相福/鄭素英 (南韓) Liu Jun (CHN) Huang Hua (CHN) Lee Sang Bok/Shon Jin Hwan Hwang Hye Young/Gil Young Ah Lee Sang Bok/Chung So Young (KOR) (KOR) (KOR) 1992 HK$466,000 吳文凱 (中國) 方銖賢 (南韓) 力奇/歷斯 (印尼) 農羣華/周蕾 (中國) 李相福/吉永雅 (南韓) Wu Weikai -

Badminton World Federation Bwf Handbook Ii
HANDBOOK I I LAWS OF BADMINTON | REGULATIONS BADMINTON WORLD FEDERATION BWF HANDBOOK II (Laws of Badminton & Regulations) 2010/2011 It is the duty of everyone concerned with badminton to keep themselves informed about the BWF Statutes COPYRIGHT ALL RIGHTS RESERVED Permission to reprint material in this book, either wholly or in part in any form whatsoever, must be obtained from the Badminton World Federation Updated 25 May 2010 by BADMINTON WORLD FEDERATION Stadium Badminton Kuala Lumpur Batu 3 ½ , Jalan Cheras 56000 Kuala Lumpur, Malaysia Tel: +603-9283 7155 / 6155 / 2155 Fax: +603-9284 7155 E-Mail: [email protected] Web: www.bwfbadminton.org CONTENTS 2 Laws of Badminton ....................................................................................................... 4 Laws of Badminton (Appendix 1-6) ............................................................................ 14 Recommendation to Technical Officials ..................................................................... 30 General Competition Regulations ............................................................................... 41 Appendix 1 - International Representation Chart ..................................................... 80 Appendix 2 - Specifications for International Standard Facilities ........................... 81 Appendix 3 – Anti Doping Regulations ....................................................................... 83 Appendix 4 – Players’ Code of Conduct ..................................................................... 120 Appendix -

Selected Results of World Badminton Championships 08:17, August 16, 2007
Selected results of world badminton championships 08:17, August 16, 2007 Following are the selected results of the 16th world badminton championships on Wednesday: Men's singles: Park Sung Hwan, South Korea, bt Andrew Dabeka, Canada, 21-16, 21-5 Kenneth Jonassen, Denmark, bt Nicholas Kidd, England, 21-14, 21-7 Chen Yu, China, bt Bjoern Joppien, Germany, 21-15, 21-7 Ronald Susilo, Singapore, bt Richard Vaughan, Wales, 21-17, 21-17 Anup Sridhar, India, bt Taufik Hidayat, Indonesia, 21-14, 24-26, 22-20 Peter Gade, Denmark, bt Chan Yan Kit, Hong Kong, China, 21-18, 25-23 Muhd Hafiz B Hashim, Malaysia, bt Scott Evans, Ireland, 21-19, 14-21, 21-11 Lee Chong Wei, Malaysia, bt Eric Pang, Netherlands, 21-7, 21-11 Lin Dan, China, bt Wei Ng, Hong Kong, China, 21-8, 21-10 Bao Chunlai, China, bt Lee Tsuen Seng, Malaysia, 21-14, 18-21, 21-18 Women's singles: Pi Hongyan, France, bt Charmaine Reid, Canada, 21-13, 21-12 Wang Chen, Hong Kong, China, bt Li Li, Singapore, 21-13, 21-6 Zhang Ning, China, bt Jang Soo Young, South Korea, 21-9, 21-14 Tracey Hallam, England, bt Kamila Augustyn, Poland, 15-21, 21-16, 21-11 Wong Mew Choo, Malaysia, bt Ekaterina Ananina, Russia, 21-14, 22-20 Xie Xingfang, China, bt Ana Moura, Portugal, 21-2, 21-7 Ella Karachkova, Russia, bt Yao Jie, Netherlands, 22-20, 14-21, 21-17 Zhu Lin, China, bt Anna Rice, Canada, 21-18, 21-13 Maria Kristin Yulianti, Indonesia, bt Wong Pei Xian Julia, Malaysia, 16-21, 21-14, 21-18 Lu Lan, China, bt Judith Meulendijks, Netherlands, 21-18, 21-13 Men's doubles: Choong Tan Fook/Lee Wan Wah, Malaysia, -

Womens Doubles All England 1899 to 2009
WOMENS DOUBLES 1899 to 2009 1899 - Muriel Lucas - later Adams/Graeme (England) 1900 - Muriel Lucas - later Adams/Graeme (England) 1901 - Daisey St.John/E. Moseley - later Allen (England) 1902 - Ethel W. Thomson - later Larcombe/Muriel Lucas - later Adams (England) 1903 - M. Hardy/Dorothea K. Douglas - later Lambert Chambers (England) 1904 - Ethel W. Thomson - later Larcombe/Muriel Lucas - later Adams (England) 1905 - Ethel W. Thomson - later Larcombe/Muriel Lucas - later Adams (England) 1906 - Ethel W. Thomson - later Larcombe/Muriel Lucas - later Adams (England) 1907 - G.L. Murray/Muriel Lucas - later Adams (England) 1908 - G.L. Murray/Muriel Lucas - later Adams (England) 1909 - G.L. Murray/Muriel Lucas - later Adams (England) 1910 - M.K. Bateman - later Flaxman/Muriel Lucas - later Adams (England) 1911 - A. Gowenlock/D. Cundall - later Bisgood (England) 1912 - A. Gowenlock/D. Cundall - later Bisgood (England) 1913 - Hazel Hogarth/M.K. Bateman - later Flaxman (England) 1914 - Margaret Rivers Tragett - formerly Larminie/Eveline Grace Peterson (England) 1915-1919 - Cancelled during World War I 1920 - Lavinia C. Radeglia/Violet Elton (England) 1921 - Kathleen 'Kitty' McKane - later Godfree/Margaret McKane - later Stocks (England) 1922 - Margaret Rivers Tragett - formerly Larminie/Hazel Hogarth (England) 1923 - Margaret Rivers Tragett - formerly Larminie/Hazel Hogarth (England) 1924 - Margaret Stocks - formerly McKane/Kathleen 'Kitty' McKane - later Godfrey (England) 1925 - Margaret Rivers Tragett - formerly Larminie/Hazel Hogarth (England) -

All England Badminton Championships Winners
All England Badminton Championships Winners Men’s Singles Year Winner 1899 No competition 1900 Sidney Howard Smith (England) 1901 Capt. H.W. Davies (England) 1902 Ralph George Watling (England) 1903 Ralph George Watling (England) 1904 Henry Norman Marrett (England) 1905 Henry Norman Marrett (England) 1906 Norman Wood (England) 1907 Norman Wood (England) 1908 Henry Norman Marrett (England) 1909 Frank Chesterton (England) 1910 Frank Chesterton (England) 1911 Guy Sautter (England) 1912 Frank Chesterton (England) 1913 Guy Sautter (England) 1914 Guy Sautter (England) 1915-19 The All England cancelled during World War I 1921 Sir George Thomas (England) 1922 Sir George Thomas (England) 1923 Sir George Thomas (England) 1924 Gordon 'Curly' Mack (Ireland) 1925 Frank Devlin (Ireland) 1926 Frank Devlin (Ireland) 1927 Frank Devlin (Ireland) 1928 Frank Devlin (Ireland) 1929 Frank Devlin (Ireland) 1930 Donald Hume (England) 1931 Frank Devlin (Ireland) 1932 Ralph Nichols (England) 1933 Raymond 'Bill' White (England) 1934 Ralph Nichols (England) 1935 Raymond 'Bill' White (England) 1936 Ralph Nichols (England) 1937 Ralph Nichols (England) 1938 Ralph Nichols (England) 1939 Tage Madsen (Denmark) 1940-46 The All England cancelled during World War II 1947 Conny Jepsen (Sweden) 1948 Jørn Skaarup (Denmark) 1949 Dave Freeman (USA) 1950 Wong Peng Soon (Malaysia) 1951 Wong Peng Soon (Malaysia) 1952 Wong Peng Soon (Malaysia) 1953 Eddy Choong (Malaysia) 1954 Eddy Choong (Malaysia) 1955 Wong Peng Soon (Malaysia) 1956 Eddy Choong (Malaysia) 1957 Eddy Choong (Malaysia) -

All England Badminton Championships Winners
All England Badminton Championships Winners Men’s Singles Year Winner 1899 No competition 1900 Sidney Howard Smith (England) 1901 Capt. H.W. Davies (England) 1902 Ralph George Watling (England) 1903 Ralph George Watling (England) 1904 Henry Norman Marrett (England) 1905 Henry Norman Marrett (England) 1906 Norman Wood (England) 1907 Norman Wood (England) 1908 Henry Norman Marrett (England) 1909 Frank Chesterton (England) 1910 Frank Chesterton (England) 1911 Guy Sautter (England) 1912 Frank Chesterton (England) 1913 Guy Sautter (England) 1914 Guy Sautter (England) 1915-19 The All England cancelled during World War I 1921 Sir George Thomas (England) 1922 Sir George Thomas (England) 1923 Sir George Thomas (England) 1924 Gordon 'Curly' Mack (Ireland) 1925 Frank Devlin (Ireland) 1926 Frank Devlin (Ireland) 1927 Frank Devlin (Ireland) 1928 Frank Devlin (Ireland) 1929 Frank Devlin (Ireland) 1930 Donald Hume (England) 1931 Frank Devlin (Ireland) 1932 Ralph Nichols (England) 1933 Raymond 'Bill' White (England) 1934 Ralph Nichols (England) 1935 Raymond 'Bill' White (England) 1936 Ralph Nichols (England) 1937 Ralph Nichols (England) 1938 Ralph Nichols (England) 1939 Tage Madsen (Denmark) 1940-46 The All England cancelled during World War II 1947 Conny Jepsen (Sweden) 1948 Jørn Skaarup (Denmark) 1949 Dave Freeman (USA) 1950 Wong Peng Soon (Malaysia) 1951 Wong Peng Soon (Malaysia) 1952 Wong Peng Soon (Malaysia) 1953 Eddy Choong (Malaysia) 1954 Eddy Choong (Malaysia) 1955 Wong Peng Soon (Malaysia) 1956 Eddy Choong (Malaysia) 1957 Eddy Choong (Malaysia)