Insights Into Chloroplast Genome Variation Across Opuntioideae (Cactaceae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
The Flora of Chad: a Checklist and Brief Analysis 1 Doi: 10.3897/Phytokeys.23.4752 Research Article Launched to Accelerate Biodiversity Research
A peer-reviewed open-access journal PhytoKeys 23: 1–17 (2013) The Flora of Chad: a checklist and brief analysis 1 doi: 10.3897/phytokeys.23.4752 RESEARCH ARTICLE www.phytokeys.com Launched to accelerate biodiversity research The Flora of Chad: a checklist and brief analysis Giuseppe Brundu1, Ignazio Camarda1 1 Department of Science for Nature and Environmental Resources (DIPNET), University of Sassari, Via Piandanna 4, 07100 Sassari, Italy Corresponding author: Giuseppe Brundu ([email protected]) Academic editor: Sandra Knapp | Received 23 January 2013 | Accepted 9 May 2013 | Published 13 May 2013 Citation: Brundu G, Camarda I (2013) The Flora of Chad: a checklist and brief analysis. PhytoKeys 23: 1–17. doi: 10.3897/phytokeys.23.4752 Abstract A checklist of the flora of Chad has been compiled by the authors, based on literature, on-line data-bases, herbarium collections and land surveys (1998-2011). It counts 2,460 records, i.e. 2,288 species (including 128 autonyms), 83 subspecies, 81 varieties, 8 forms, while all the previous available information reported 1,600 species. They belong to 151 Families, with 48.7% of the taxa belonging to the 6 largest families, i.e. Poaceae (14.6%), Fabaceae (13.6%), Cyperaceae (7.0%), Asteraceae (6.2 %), Malvaceae (3.9%) and Rubiaceae (3.4%). A total number or 2,173 species (88.3%) are native to Chad, including 55 (2.2%) endemic species, while 274 (11.0%) are alien to Chad, and 13 (0.5%) are considered cryptogenic, i.e. of uncertain status. It represents a considerable update on previous knowledge on the alien flora of Chad that counted for 131 taxa (5.3%). -
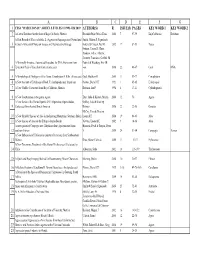
Haseltonia Articles and Authors.Xlsx
ABCDEFG 1 CSSA "HASELTONIA" ARTICLE TITLES #1 1993–#26 2019 AUTHOR(S) R ISSUE(S) PAGES KEY WORD 1 KEY WORD 2 2 A Cactus Database for the State of Baja California, Mexico Resendiz Ruiz, María Elena 2000 7 97-99 BajaCalifornia Database A First Record of Yucca aloifolia L. (Agavaceae/Asparagaceae) Naturalized Smith, Gideon F, Figueiredo, 3 in South Africa with Notes on its uses and Reproductive Biology Estrela & Crouch, Neil R 2012 17 87-93 Yucca Fotinos, Tonya D, Clase, Teodoro, Veloz, Alberto, Jimenez, Francisco, Griffith, M A Minimally Invasive, Automated Procedure for DNA Extraction from Patrick & Wettberg, Eric JB 4 Epidermal Peels of Succulent Cacti (Cactaceae) von 2016 22 46-47 Cacti DNA 5 A Morphological Phylogeny of the Genus Conophytum N.E.Br. (Aizoaceae) Opel, Matthew R 2005 11 53-77 Conophytum 6 A New Account of Echidnopsis Hook. F. (Asclepiadaceae: Stapeliae) Plowes, Darrel CH 1993 1 65-85 Echidnopsis 7 A New Cholla (Cactaceae) from Baja California, Mexico Rebman, Jon P 1998 6 17-21 Cylindropuntia 8 A New Combination in the genus Agave Etter, Julia & Kristen, Martin 2006 12 70 Agave A New Series of the Genus Opuntia Mill. (Opuntieae, Opuntioideae, Oakley, Luis & Kiesling, 9 Cactaceae) from Austral South America Roberto 2016 22 22-30 Opuntia McCoy, Tom & Newton, 10 A New Shrubby Species of Aloe in the Imatong Mountains, Southern Sudan Leonard E 2014 19 64-65 Aloe 11 A New Species of Aloe on the Ethiopia-Sudan Border Newton, Leonard E 2002 9 14-16 Aloe A new species of Ceropegia sect. -

Useful Concepts
Useful Concepts An accidental bonsai (2018). This is a Genovese basil plant, a Western kitchen herb. It’s not supposed to be very long-lived. This is an old plant, and the trunk looks terrible. It has split and the bark is flaking. Some stems are also dead. Likewise, for cacti that are short- lived, you will be hard-pressed to keep them in great shape for many years. Many cacti are just green stems – how many regular green-stemmed leafy plants do you know that can live for many, many years and stay in good shape? The following piece is part of a collection of writings published on the Practical Small Cacti Malaysia site . Things to Think About Writing is a really useful activity, especially when it comes to writing about the cultivation of cacti in a tropical climate. Many a time, I thought I have understood something, but when I went over the material after a few months, I realized that I was on the wrong path. Over time, I have taken note of a number of concepts – things to think about – that helped to direct my lines of thought in the right direction. If we understand cactus plants better, we can take better care of them. -1- Nicknames for Scientific Names PMag = Parodia magnifica GBald = Gymnocalycium baldianum PClav = Parodia claviceps MGeo = Myrtillocactus geometrizans This naming scheme is purely for convenience. Just think of them as webchat nicknames. It’s All Kinds of Green Stems Some MGeo specimens, January 2019. Growing cacti means dealing with all kinds of green stems. -

Estudios Citogenéticos Y De Contenido De ADN En Brasiliopuntia Schulzii (Cactaceae)
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by CONICET Digital Gayana Bot. 73(2):73(2), 2016414-420, 2016 ISSN 0016-5301 Estudios citogenéticos y de contenido de ADN en Brasiliopuntia schulzii (Cactaceae) Cytogenetic studies and DNA content in Brasiliopuntia schulzii (Cactaceae) MARÍA LAURA LAS PEÑAS1*, FEDERICO SANTIÑAQUE2, BEATRIZ LÓPEZ-CARRO2 & LAURA STIEFKENS1 1Instituto Multidisciplinario de Biología Vegetal (UNC- CONICET), C. C. 495 Córdoba, Argentina. 2Instituto de Investigaciones Biológicas Clemente Estable, Avenida Italia 3318. CP. 11600, Montevideo, Uruguay. *[email protected] RESUMEN Se determinaron número cromosómico, cariotipo, patrones de bandeo, localización de genes ribosómicos y contenido de ADN nuclear, en dos poblaciones de Brasiliopuntia schulzii (Cactaceae, subfam. Opuntioideae). La especie resultó diploide (2n=22), presentó una fórmula cariotípica de 10 m + 1 sm. El bandeo cromosómico CMA/DAPI reveló la presencia de un par cromosómico con una banda CMA+/DAPI- asociada a NORs. Con la técnica de FISH se observó que los loci 18- 5,8-26S fueron consistentes con los bloques CMA+/DAPI-/NORs. La señal para el gen 5S se localizó en el cuarto par m en una región telomérica. En las dos poblaciones el análisis de contenido de ADN reveló una mezcla de núcleos con tres picos de 2C, 4C y 8C. Esto indicaría un proceso de endopoliploidía en la especie. Los resultados reportados en este trabajo, combinados con las características morfológicas, indicarían que este género posee caracteres basales, concordando con los análisis filogenéticos de la subfamilia. PALABRAS CLAVE: Brasiliopuntia, cariotipo, contenido de ADN, endopoliploidía, heterocromatina, genes ribosomales. -

Caryophyllales 2018 Instituto De Biología, UNAM September 17-23
Caryophyllales 2018 Instituto de Biología, UNAM September 17-23 LOCAL ORGANIZERS Hilda Flores-Olvera, Salvador Arias and Helga Ochoterena, IBUNAM ORGANIZING COMMITTEE Walter G. Berendsohn and Sabine von Mering, BGBM, Berlin, Germany Patricia Hernández-Ledesma, INECOL-Unidad Pátzcuaro, México Gilberto Ocampo, Universidad Autónoma de Aguascalientes, México Ivonne Sánchez del Pino, CICY, Centro de Investigación Científica de Yucatán, Mérida, Yucatán, México SCIENTIFIC COMMITTEE Thomas Borsch, BGBM, Germany Fernando O. Zuloaga, Instituto de Botánica Darwinion, Argentina Victor Sánchez Cordero, IBUNAM, México Cornelia Klak, Bolus Herbarium, Department of Biological Sciences, University of Cape Town, South Africa Hossein Akhani, Department of Plant Sciences, School of Biology, College of Science, University of Tehran, Iran Alexander P. Sukhorukov, Moscow State University, Russia Michael J. Moore, Oberlin College, USA Compilation: Helga Ochoterena / Graphic Design: Julio C. Montero, Diana Martínez GENERAL PROGRAM . 4 MONDAY Monday’s Program . 7 Monday’s Abstracts . 9 TUESDAY Tuesday ‘s Program . 16 Tuesday’s Abstracts . 19 WEDNESDAY Wednesday’s Program . 32 Wednesday’s Abstracs . 35 POSTERS Posters’ Abstracts . 47 WORKSHOPS Workshop 1 . 61 Workshop 2 . 62 PARTICIPANTS . 63 GENERAL INFORMATION . 66 4 Caryophyllales 2018 Caryophyllales General program Monday 17 Tuesday 18 Wednesday 19 Thursday 20 Friday 21 Saturday 22 Sunday 23 Workshop 1 Workshop 2 9:00-10:00 Key note talks Walter G. Michael J. Moore, Berendsohn, Sabine Ya Yang, Diego F. Registration -

What Did the First Cacti Look Like
What Did the First Cactus Look like? An Attempt to Reconcile the Morphological and Molecular Evidence Author(s): M. Patrick Griffith Source: Taxon, Vol. 53, No. 2 (May, 2004), pp. 493-499 Published by: International Association for Plant Taxonomy (IAPT) Stable URL: http://www.jstor.org/stable/4135628 . Accessed: 03/12/2014 10:33 Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at . http://www.jstor.org/page/info/about/policies/terms.jsp . JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. International Association for Plant Taxonomy (IAPT) is collaborating with JSTOR to digitize, preserve and extend access to Taxon. http://www.jstor.org This content downloaded from 192.135.179.249 on Wed, 3 Dec 2014 10:33:44 AM All use subject to JSTOR Terms and Conditions TAXON 53 (2) ' May 2004: 493-499 Griffith * The first cactus What did the first cactus look like? An attempt to reconcile the morpholog- ical and molecular evidence M. Patrick Griffith Rancho Santa Ana Botanic Garden, 1500 N. College Avenue, Claremont, California 91711, U.S.A. michael.patrick. [email protected] THE EXTANT DIVERSITYOF CAC- EARLYHYPOTHESES ON CACTUS TUS FORM EVOLUTION Cacti have fascinated students of naturalhistory for To estimate evolutionaryrelationships many authors many millennia. Evidence exists for use of cacti as food, determinewhich morphological features are primitive or medicine, and ornamentalplants by peoples of the New ancestral versus advanced or derived. -

Universidad Nacional De San Agustín De Arequipa
UNIVERSIDAD NACIONAL DE SAN AGUSTÍN DE AREQUIPA ESCUELA DE POSGRADO UNIDAD DE POSGRADO DE LA FACULTAD DE CIENCIAS NATURALES Y FORMALES “IDENTIFICACIÓN Y ESTADO DE CONSERVACIÓN DE CACTÁCEAS EN PUERTO CORIO, DISTRITO DE PUNTA DE BOMBON, PROVINCIA DE ISLAY Y PROPUESTA DE CONSERVACION 2018” Tesis presentado por el Maestro JOSE ELIAS ZUÑIGA CALCINA Para optar el Grado Académico de Doctor en Ciencias y Tecnologías medioambientales Asesora: Trinidad Betty Paredes de Gómez AREQUIPA – PERÚ 2019 ii “IDENTIFICACIÓN Y ESTADO DE CONSERVACIÓN DE CACTACEAS EN PUERTO CORIO, DISTRITO DE PUNTA DE BOMBON, PROVINCIA DE ISLAY Y PROPUESTA DE CONSERVACIÓN 2018” Tesis presentada por: JOSÉ ELIAS ZÚÑIGA CALCINA Jurado: Dra. MARIA ELEANA VARGAS DE NIETO ______________________ Dr. PAUL KAREL HUANCA ZUÑIGA ______________________ Dra. TRINIDAD BETTY PAREDES DE GOMEZ ______________________ iii DEDICATORIA A mis padres, esposa e hijas, con mucho amor y cariño, les dedico todo mi esfuerzo y trabajo puesto en la realización de la presente tesis. iv CONTENIDO Página RESUMEN ABSTRACT CAPITULO I: FUNDAMENTACIÓN DE LA INVESTIGACIÓN 01 1.1. Problema 01 1.2. Estado de Arte 02 1.3. Justificación 05 1.4. Objetivo General 06 1.5. Objetivos Específicos 06 1.6. Hipótesis 06 CAPITULO II: MARCO TEÓRICO 07 2.1. Biodiversidad 07 2.2. El cambio climático 08 2.3. Endemismo 10 2.4. Estado de Conservación 11 2.5. Las Cactáceas 13 2.5.1. Origen de las Cactáceas 14 2.5.2. Diversidad de cactáceas en el continente americano 15 2.5.3. Interacción con otras Plantas y Animales de un Ecosistema 16 2.5.3.1.Polinización 16 2.5.3.2.Dispersión de Semillas 16 2.5.4. -

Cactus (Opuntia Spp.) As Forage 169
Cactus (Opuntia spp.) as forage 169 Food •••A.gricultv,.. Org•nU.taon or United -N••lon• FAO Cactus (Opuntiaspp.) PLANT PRODUCTION as forage AND PROTECTlON PAPER 169 Ed~ed by Candelario Mondragon-Jacobo lnstituto Nacional de Investigaciones Forestales y Agropecuarias (INIFAP) Mexico and Salvador Perez-Gonzalez Universidad Aut6noma de Queretaro Mexico Coordinated for FAD by Enrique Arias Horticultural Crops Group Stephen G. Reynolds Grassland and Pasture Crops Group FAO Plant Production and Protection Division and Manuel D. sanchez Feed Resources Group FAO Animal Production and HeaHh Division Produced within the frameworl< of the FAO International Technical Cooperation Networl< ot on Cactus Pear ••u nttttd• NaUon• Rome,2001 Reprinted 2002 The designations “developed” and “developing” economies are intended for statistical convenience and do not necessarily express a judgement about the stage reached by a particular country, country territory or area in the development process. The views expressed herein are those of the authors and do not necessarily represent those of the Food and Agriculture Organization of the United Nations or of their affiliated organization(s). The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. ISBN 92-5-104705-7 All rights reserved. Reproduction and dissemination of material in this information product for educational or other non-commercial purposes are authorized without any prior written permission from the copyright holders provided the source is fully acknowledged. -

The Natural World That I Seek out in the Desert Regions of Baja California
The natural world that I seek out in the desert regions of Baja California and southern California provides me with scientific adventure, excitement towards botany, respect for nature, and overall feelings of peace and purpose. Jon P. Rebman, Ph.D. has been the Mary and Dallas Clark Endowed Chair/Curator of Botany at the San Diego Natural History Museum (SDNHM) since 1996. He has a Ph.D. in Botany (plant taxonomy), M.S. in Biology (floristics) and B.S. in Biology. Dr. Rebman is a plant taxonomist and conducts extensive floristic research in Baja California and in San Diego and Imperial Counties. He has over 15 years of experience in the floristics of San Diego and Imperial Counties and 21 years experience studying the plants of the Baja California peninsula. He leads various field classes and botanical expeditions each year and is actively naming new plant species from our region. His primary research interests have centered on the systematics of the Cactus family in Baja California, especially the genera Cylindropuntia (chollas) and Opuntia (prickly-pears). However, Dr. Rebman also does a lot of general floristic research and he co- published the most recent edition of the Checklist of the Vascular Plants of San Diego County. He has over 22 years of field experience with surveying and documenting plants including rare and endangered species. As a field botanist, he is a very active collector of scientific specimens with his personal collections numbering over 22,500. Since 1996, he has been providing plant specimen identification/verification for various biological consulting companies on contracts dealing with plant inventory projects and environmental assessments throughout southern California. -

Cactaceae) Ve Výuce Biologie Na Středních Školách
UNIVERZITA PALACKÉHO V OLOMOUCI PŘÍRODOVĚDĚCKÁ FAKULTA KATEDRA BOTANIKY Čeleď kaktusovité (Cactaceae) ve výuce biologie na středních školách DIPLOMOVÁ PRÁCE Bc. Adéla Gorová Biologie N1501, Biologie – Geografie Prezenční studium Vedoucí práce: Mgr. Martina Oulehlová, Ph.D. Olomouc 2020 Prohlášení Prohlašuji, že předložená práce je mým původním autorským dílem, které jsem vypracovala samostatně. Veškerou literaturu a další zdroje, z nichž jsem při zpracování čerpala, v práci řádně cituji a jsou uvedeny v seznamu použité literatury. V Olomouci dne . Adéla Gorová Poděkování Mé poděkování patří vedoucí bakalářské práce Mgr. Martině Oulehlové, Ph. D. za odborné vedení, ochotu a čas, který mi v průběhu vypracovávání diplomové práce věnovala. Dále poděkování patří Ing. Heleně Šupové, Ing. Zdeňku Šupovi a Ing. Pavlu Součkovi za poskytnutí materiálů a umožnění vstupu do Kaktusového skleníku Výstaviště Flora Olomouc, a.s., a také Nikol Kaletové za odbornou korekci abstraktu přeloženého do angličtiny. Poděkování patří také projektům IGA-Prf- 2018-001 a IGA-Prf-2019-004. BIBLIOGRAFICKÁ IDENTIFIKACE Jméno a příjmení: Bc. Adéla Gorová Název práce: Čeleď kaktusovité (Cactaceae) ve výuce biologie na středních školách Typ práce: Diplomová práce Pracoviště: Katedra botaniky, Přírodovědecká fakulta, Univerzita Palackého v Olomouci Vedoucí práce: Mgr. Martina Oulehlová, Ph.D. Rok obhajoby: 2020 Abstrakt: Diplomová práce se zabývá problematikou výuky čeledi kaktusovitých (Cactaceae) na středních školách. Teoretická část práce je zaměřena na praktický význam, využití, zajímavosti a specifika čeledi Cactaceae. Dále na charakteristiku Kaktusového skleníku Výstaviště Flora Olomouc, a.s., rozmístění zástupců kaktusů ve skleníku a charakteristiku pěstovaných zástupců kaktusů. Praktická část je zaměřena na tvorbu přehledného systému čeledi Cactaceae pro výuku studentů na středních školách, na přípravu přehledu pěstovaných zástupců kaktusů a na vytvoření komplexní přípravy pro realizaci exkurze pedagoga se studenty do sbírkového Kaktusového skleníku Výstaviště Flora Olomouc, a.s. -

Projeto De Pesquisa
INSTITUTO DE PESQUISAS JARDIM BOTÂNICO DO RIO DE JANEIRO DIRETORIA DE PESQUISA CIENTÍFICA PROGRAMA INSTITUCIONAL DE BOLSAS DE INICIAÇÃO CIENTÍFICA (PIBIC/CNPq) PROJETO DE PESQUISA ESTUDOS MULTIDISCIPLINARES EM MELASTOMATACEAE NEOTROPICAIS: TAXONOMIA, FLORÍSTICA, ANATOMIA E ONTOGENIA Dr. José Fernando A. Baumgratz (orientador proponente) PLANOS DE TRABALHO (1) VASCULARIZAÇÃO E ONTOGENIA DE FLORES POLISTÊMONES EM MICONIA (MELASTOMATACEAE; MICONIEAE) (2) DIVERSIDADE DE MELASTOMATACEAE NO PARQUE NACIONAL DA SERRA DOS ÓRGÃOS, RIO DE JANEIRO, BRASIL Rio de Janeiro 2018 1 PROJETO DE PESQUISA ESTUDOS MULTIDISCIPLINARES EM MELASTOMATACEAE NEOTROPICAIS: TAXONOMIA, FLORÍSTICA E ANATOMIA INTRODUÇÃO A família Melastomataceae é uma das mais numerosas entre as Angiospermae, com 150 gêneros e em torno de 4.500 espécies (Renner et al. 2010), sendo floristicamente abundante e diversificada na América do Sul. A expressiva diversidade dessa família na flora brasileira está seguramente registrada na lista de plantas do Brasil recentemente divulgada, onde vários taxonomistas especialistas atestaram a validade dos táxons que a compõe (Baumgratz et al. 2010). No contexto nacional, representa a sexta maior família entre as Angiospermae, com 68 gêneros e mais de 1.300 espécies, sendo 845 endêmicas, e distribuindo-se desde a Amazônia e o centro-oeste até o Rio Grande do Sul e praticamente em todas as formações vegetacionais, exceto na Caatinga sensu stricto (Baumgratz & Souza 2005; Baumgratz et al. 2006, 2007). Entretanto, de acordo com Goldenberg et al. (2012), o número de gêneros poderá cair para 65, com base em estudos filogenéticos recentemente desenvolvidos, como os de Penneys et al. (2010) e Penneys & Judd (2011), que predizem alguns como sinônimos. De modo geral, a base do conhecimento sobre as Melastomataceae no Brasil ainda são as clássicas monografias publicadas no final do século XIX por Cogniaux (1883- 1888, 1891). -

Universidade Estadual De Campinas Instituto De
UNIVERSIDADE ESTADUAL DE CAMPINAS INSTITUTO DE BIOLOGIA JULIA MEIRELLES FILOGENIA DE Miconia SEÇÃO Miconia SUBSEÇÃO Seriatiflorae E REVISÃO TAXONÔMICA DO CLADO ALBICANS (MELASTOMATACEAE, MICONIEAE) PHYLOGENY OF Miconia SECTION Miconia SUBSECTION Seriatiflorae AND TAXONOMIC REVIEW OF THE ALBICANS CLADE (MELASTOMATACEAE, MICONIEAE) CAMPINAS 2015 UNIVERSIDADE ESTADUAL DE CAMPINAS INSTITUTO DE BIOLOGIA Dedico ao botânico mais importante da minha vida: Leléu. Por tudo. AGRADECIMENTOS Agradeço formalmente as agências que concederam as bolsas de estudos sem as quais seria impossível o desenvolvimento deste trabalho ao longo dos últimos 4 anos e meio: CAPES (PNADB e PDSE), CNPQ (programa REFLORA) e NSF (EUA) no âmbito do projeto PBI Miconieae por financiar o trabalho em campo e herbários na Amazônia e também a parte molecular apresentada no Capítulo I. Foram tantas as pessoas que me acompanharam nessa jornada ou que felizmente cruzaram o meu caminho ao decorrer dela, que não posso chegar ao destino sem agradecê-las... O meu maior agradecimento vai ao meu orientador sempre presente Dr. Renato Goldenberg que desde o mestrado confiou em meu trabalho e compartilhou muito conhecimento sobre taxonomia, Melastomataceae e às vezes, até mesmo vida. Sem o seu trabalho, seu profissionalismo e a sua paciência esta tese jamais seria possível. Agradeço pelo tempo que dedicou em me ajudar tanto a crescer como profissional e também como pessoa. Agradeço muito ao meu co-orientador Dr. Fabián Michelangeli por todos os ensinamentos e ajudas no período de estágio sanduíche no Jardim Botânico de Nova Iorque. Todo o seu esforço em reunir e compartilhar bibliografias, angariar recursos e treinar alunos (no quais eu me incluo) tem feito toda a diferença na compreensão da sistemática de Melastomataceae e deixado frutos de inestimável valor.