Testing the Utility of Partial COI Sequences for Phylogenetic Estimates of Gastropod Relationships
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metabolic Response of Antarctic Pteropods (Mollusca: Gastropoda) to Food Deprivation and Regional Productivity
Vol. 441: 129–139, 2011 MARINE ECOLOGY PROGRESS SERIES Published November 15 doi: 10.3354/meps09358 Mar Ecol Prog Ser Metabolic response of Antarctic pteropods (Mollusca: Gastropoda) to food deprivation and regional productivity Amy E. Maas1,*, Leanne E. Elder1, Heidi M. Dierssen2, Brad A. Seibel1 1Department of Biological Sciences, University of Rhode Island, Kingston, Rhode Island 02881, USA 2Department of Marine Sciences, University of Connecticut, Groton, Connecticut 06340, USA ABSTRACT: Pteropods are an abundant group of pelagic gastropods that, although temporally and spatially patchy in the Southern Ocean, can play an important role in food webs and biochem- ical cycles. We found that the metabolic rate in Limacina helicina antarctica is depressed (~23%) at lower mean chlorophyll a (chl a) concentrations in the Ross Sea. To assess the specific impact of food deprivation on these animals, we quantified aerobic respiration and ammonia (NH3) produc- tion for 2 dominant Antarctic pteropods, L. helicina antarctica and Clione limacina antarctica. Pteropods collected from sites west of Ross Island, Antarctica were held in captivity for a period of 1 to 13 d to determine their metabolic response to laboratory-induced food deprivation. L. helicina antarctica reduced oxygen consumption by ~20% after 4 d in captivity. Ammonia excretion was not significantly affected, suggesting a greater reliance on protein as a substrate for cellular res- piration during starvation. The oxygen consumption rate of the gymnosome, C. li macina antarc- tica, was reduced by ~35% and NH3 excretion by ~55% after 4 d without prey. Our results indi- cate that there is a link between the large scale chl a concentrations of the Ross Sea and the baseline metabolic rate of pteropods which impacts these animals across multiple seasons. -

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Validation of Holoplanktonic Molluscan Taxa from the Oligo- Miocene of the Maltese Archipelago, Introduced in Violation with ICZN Regulations
Cainozoic Research, 9(2), pp. 189-191, December 2012 ! ! ! Validation of holoplanktonic molluscan taxa from the Oligo- Miocene of the Maltese Archipelago, introduced in violation with ICZN regulations Arie W. Janssen Naturalis Biodiversity Center, P.O. Box 9517, 2300 RA Leiden, The Netherlands; currently: 12 Triq tal’Hamrija, Xewkija XWK 9033, Gozo, Malta; email: [email protected] Received 29 August 2012, accepted 6 September 2012 Five gymnosomatous molluscan taxa were recently introduced applying ‘open generic nomenclature’ by using the indication ‘Genus Clionidarum’ instead of a formal genus name and therefore violating ICZN art. 11.9.3 of the Code. Herein those taxa are validated by placing them in the type genus of the family Clionidae, followed by a question mark indicating here that they might as well belong to any other of the known (or as yet unknown) genera in the family Clionidae . Introduction Clionidarum’ as used in my paper indeed cannot be con- sidered to be an unambiguous genus name. In my study of Maltese fossil holoplanktonic molluscs I herewith validate the new names by combining them with (Janssen, 2012) a number of new species of gymnosoma- the unambiguous genus name Clione, followed by a ques- tous larval shells were introduced, more or less resembling tion mark, indicating here that those species might as well the few Recent larval shells known from this group of Gas- belong to any other known or unknown genus in the tropoda, but obviously (also considering their ages) repre- Clionidae. senting undescribed species. Recent Gymnosomata are RGM-registration numbers refer to the collections of Natu- shell-less in the adult stage, and their larval shell is shed at ralis Biodiversity Center, Palaeontology Department (Lei- metamorphosis from larva to adult. -

Spinucella a New Genus of Miocene to Pleistocene , Muricid Gastropods from the Eastern Atlantic
Contr. Tert. Quatern. Geol. 30(1-2) 19-27 1 tab., 1 pi. Leiden, June 1993 Spinucella a new genus of Miocene to Pleistocene , muricid gastropods from the eastern Atlantic Geerat J. Vermeij University of California Davis, U.S.A. — new of Miocene Pleistocene muricid from the Atlantic. Contr. Tert. Vermeij, GeeratJ. Spinucella, a genus to gastropods eastern Quatern. Geol., 30(1-2): 19-27, 1 tab., 1 pi. Leiden, June 1993. The muricid is for de C. 1825 from the Pliocene of the new gastropod genus Spinucella proposed Purpura tetragonaJ. Sowerby, (type species), North Sea Basin, and for several other early Miocene to late Pleistocene species from southern Europe, North Africa, and southern Africa. The is characterised the of labral on the of the shell and reticulate of genus by presence a spine outer lip by sculpture. Species and Acanthinucella Cooke, 1918. The Spinucella closely resemble members ofNucella Röding, 1798, Acanthina Fischer von Waldheim, 1807, ofthat in the eastern Pacific Acanthina andAcanthinucella. Withthe removal of labral spine of Spinucellawas probably evolved independendy from where authors have the the time of arrival of Nucella in the North Atlantic from the S. tetragona Nucella, many recent placed species, North Pacific was late Pliocene, rather than middle Pliocene. — words Key Spinucella, new genus, Acanthina, Nucella, Neogene, biogeography. Prof. Dr G.J. Vermeij, Department of Geology, University of California, Davis, CA 95616, U.S.A. Contents 1956; Glibert, 1959, 1963). In fact, Glibert (1959) considered N. tetragona to be ancestral to N. lapillus, Introduction 19 p. the two species being linked by the late Pliocene 20 Systematics p. -

Gyraulus Laevis in Nederland 87
Kuijper: Gyraulus laevis in Nederland 87 Gyraulus laevis (Mollusca: Planorbidae) in Nederland door W.J. Kuijper Enkele recente waarnemingen van een van onze zeldzaamste planorbiden waren de aanleiding tot het samenstellen van een over- laevis zicht van alle van Nederland bekende vondsten van Gyraulus (Alder, 1838). Tot voor kort waren slechts enkele vindplaatsen van dit dier bekend (Janssen & De Vogel, 1965: 75). Hierbij komt het betrof feit dat het in een aantal gevallen slechts een enkel exemplaar en de soort niet meer teruggevonden kon worden. Het volgende geeft een chronologisch overzicht van de waarnemingen van Gyraulus laevis in Nederland. Voor zover beschikbaar zijn diverse gegevens van de vindplaatsen vermeld. WAARNEMINGEN 1. Koudekerke (bij Middelburg), buitenplaats Vijvervreugd, ± 1890. Fraai vers materiaal: 69 exemplaren in de collectie Schep- de man (Zoölogisch Museum, Amsterdam). Later niet meer vermeld, buitenplaats bestaat niet meer (Kuiper, 1944: 6). Dit materiaalwerd verzameld door Dr. IJ. Keijzer en is vermeld in: Verslag over 1885-1893 van het Zeeuwsch Genootschap der Wetenschappen, blz. 88 (volgens Mevr. Dr. W.S.S. van der Feen - van Benthem Jut- ting). Verder is materiaal van deze vindplaats in de collecties van het Zeeuwsch Museum te Middelburg (2 ex.) en het Natuurhistorisch Museum te Enschede (6 ex.) aanwezig. 2. Warmond (bij Leiden), in de Leede, januari 1916 (Van Ben- them Jutting, 1933: 179). Dit materiaal werd verzameld door P.P. de Koning; in het Rijksmuseum van Natuurlijke Historie te Leiden bevinden zich vier schelpen van deze vindplaats, waarvan twee een doorsnede van ca. 4 mm bereiken. 3. 't Zand (bij Roodeschool), Oostpolder, 1927 (Van Benthem Jutting, 1947: 66). -

Metacommunities and Biodiversity Patterns in Mediterranean Temporary Ponds: the Role of Pond Size, Network Connectivity and Dispersal Mode
METACOMMUNITIES AND BIODIVERSITY PATTERNS IN MEDITERRANEAN TEMPORARY PONDS: THE ROLE OF POND SIZE, NETWORK CONNECTIVITY AND DISPERSAL MODE Irene Tornero Pinilla Per citar o enllaçar aquest document: Para citar o enlazar este documento: Use this url to cite or link to this publication: http://www.tdx.cat/handle/10803/670096 http://creativecommons.org/licenses/by-nc/4.0/deed.ca Aquesta obra està subjecta a una llicència Creative Commons Reconeixement- NoComercial Esta obra está bajo una licencia Creative Commons Reconocimiento-NoComercial This work is licensed under a Creative Commons Attribution-NonCommercial licence DOCTORAL THESIS Metacommunities and biodiversity patterns in Mediterranean temporary ponds: the role of pond size, network connectivity and dispersal mode Irene Tornero Pinilla 2020 DOCTORAL THESIS Metacommunities and biodiversity patterns in Mediterranean temporary ponds: the role of pond size, network connectivity and dispersal mode IRENE TORNERO PINILLA 2020 DOCTORAL PROGRAMME IN WATER SCIENCE AND TECHNOLOGY SUPERVISED BY DR DANI BOIX MASAFRET DR STÉPHANIE GASCÓN GARCIA Thesis submitted in fulfilment of the requirements to obtain the Degree of Doctor at the University of Girona Dr Dani Boix Masafret and Dr Stéphanie Gascón Garcia, from the University of Girona, DECLARE: That the thesis entitled Metacommunities and biodiversity patterns in Mediterranean temporary ponds: the role of pond size, network connectivity and dispersal mode submitted by Irene Tornero Pinilla to obtain a doctoral degree has been completed under our supervision. In witness thereof, we hereby sign this document. Dr Dani Boix Masafret Dr Stéphanie Gascón Garcia Girona, 22nd November 2019 A mi familia Caminante, son tus huellas el camino y nada más; Caminante, no hay camino, se hace camino al andar. -
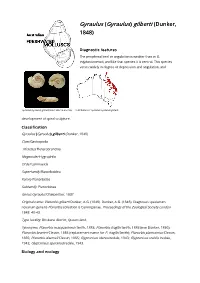
Gyraulus) Gilberti (Dunker, 1848
Gyraulus (Gyraulus) gilberti (Dunker, 1848) Diagnostic features The peripheral keel or angulation is weaker than in G. edgbastonensis, and like that species it is central. This species varies widely in degree of depression and angulation, and Gyraulus (Gyraulus) gilberti (adult size 4.8-5.5 mm) Distribution of Gyraulus (Gyraulus) gilberti. development of spiral sculpture. Classification Gyraulus (Gyraulus) gilberti (Dunker, 1848) Class Gastropoda I nfraclass Heterobranchia Megaorder Hygrophila Order Lymnaeida Superfamily Planorboidea Family Planorbidae Subfamily: Planorbinae Genus Gyraulus Charpentier, 1837 Original name: Planorbis gilberti Dunker, A.G. (1848). Dunker, A.G. (1848). Diagnoses specierum novarum generis Planorbis collection is Cumingianae. Proceedings of the Zoological Society London 1848: 40-43. Type locality: Brisbane district, Queensland. Synonyms: Planorbis macquariensis Smith, 1883; Planorbis fragilis Smith, 1883 (non Dunker, 1850); Planorbis brazieri Clessin, 1885 (replacement name for P. fragilis Smith); Planorbis planissimus Clessin, 1885; Planorbis daemeli Clessin, 1885; Glyptanisus idenusredale, 1943; Glyptanisus stabilis redale, 1943; Glyptanisus speranusredale, 1943. Biology and ecology This species lives in water weeds and other vegetation in ponds, billabongs, swamps and sluggish streams and rivers in tropical and subtropical eastern Australia. Feeds on detritus. Egg mass presumably a jelly strip containing small eggs. Development direct. Brown (2001) described the anatomy of this species. This species is an intermediate host for the stomach fluke Orthocoelium streptocoelium (Boray, 1982; Beesley et al., 1998). Distribution This species occurs throughout eastern Australia, from Cape York to northern New South Wales. Notes G. isingi and/or G.waterhousei may possibly be conspecific with this species (Brown, 2001). Further reading Beesley, P. L., Ross, G. J. -

Marine Biology 79,289-293 (1984) Marine :E:~ Biology @ Springer-Vertag 1984
Marine Biology 79,289-293 (1984) Marine :E:~ Biology @ Springer-Vertag 1984 Growth, reproduction and mortality of the sea hare Dolabella auricularia (Gastropoda: Aplysiidae) in the Central Visayas, Philippines * D.Pauly1 and H. Calumpong 2 1 International Center for LivingAquatic Resources Management (ICLARM); MCC P.O. Box 1501,Makati, Metro Manila, Philippines 2 Marine Laboratory, Silliman University; Dumaguete City, Philippines Abstract Material and methods The growth parameters WIX) and K of the von Bertalanff)r The growth equation used here to describe the growth of growth equation were estimated from size-frequency data Dolabella auricularia is the well-known von Bertalanff)r of the sea hare Dolabella auricularia using an objective, growth function (VBGF) (for weight growth): computer-based method. The results obtained from two W1= W IX)(1_e-K(t-to»3 . (1) locations in the central Philippines were comparable, although based on only a few individuals; the means for The exponent in Eq. (1) assumes that growth is isometric, both sites were WIX)=493g and K=0.9 (yearly basis). A i.e. that weight is proportional to length cubed. Since it is value of Z = 3.66, corresponding to an annual survival rate extremely difficult to take consistent linear measurements of 2.6%,was estimated for the juveniles and adults from a in aplysiid gastropods (which lack hard parts), the proce- length-converted catch curve. Spawning and recruitment dure used here was to take the cubic root of weights and were found to occur throughout the year with peaks in to treat them - under the assumption of isometry - as May to July and September to October. -

Identifying Liver Fluke Snails
Identifying liver fluke snails March 2017, Primefact 476, second edition Dr Joan Lloyd, former Veterinary Research Officer, EMAI Dr Joseph C Boray, former Principal Research Scientist, EMAI Dr Noel Campbell, former Senior Research Scientist, Department of Primary Industries, Victoria (Revised by) Stephen Love, Veterinarian/Research Officer (Parasitology), Sheep Industries, Armidale Introduction Finding liver fluke snails In NSW, about 20 million sheep and 2 million Liver fluke snails live in the mud or on plants in cattle graze pastures where liver fluke (Fasciola shallow water at the edge of springs, small hepatica) commonly occurs. Liver fluke is creeks, dam inflows and outflows, irrigation widespread across eastern NSW, where average channels, poorly drained drainage channels or in rainfall is about 600 mm or more a year. water troughs. They are small and sometimes Specifically, it occurs on the tablelands and difficult to find. nearby slopes, and the north and south coasts. It The kind of habitat in which the snail is found is also found in irrigation areas further west, often gives clues as to which type it is. For where the annual rainfall may only be 400 mm, example, Austropeplea (Lymnaea) tomentosa but is supplemented by regular irrigation. prefers trickling creeks flowing from hillside During its life cycle, liver fluke must develop in a springs and soaks (black bogs), and is only rarely particular type of small freshwater snail. found in dams, water troughs or large creeks. It can, however, be found in dam overflows after In Australia, the most important intermediate host heavy rain, or within spring-fed dam inflows and is the indigenous freshwater snail, Austropeplea outflows. -

THE LISTING of PHILIPPINE MARINE MOLLUSKS Guido T
August 2017 Guido T. Poppe A LISTING OF PHILIPPINE MARINE MOLLUSKS - V1.00 THE LISTING OF PHILIPPINE MARINE MOLLUSKS Guido T. Poppe INTRODUCTION The publication of Philippine Marine Mollusks, Volumes 1 to 4 has been a revelation to the conchological community. Apart from being the delight of collectors, the PMM started a new way of layout and publishing - followed today by many authors. Internet technology has allowed more than 50 experts worldwide to work on the collection that forms the base of the 4 PMM books. This expertise, together with modern means of identification has allowed a quality in determinations which is unique in books covering a geographical area. Our Volume 1 was published only 9 years ago: in 2008. Since that time “a lot” has changed. Finally, after almost two decades, the digital world has been embraced by the scientific community, and a new generation of young scientists appeared, well acquainted with text processors, internet communication and digital photographic skills. Museums all over the planet start putting the holotypes online – a still ongoing process – which saves taxonomists from huge confusion and “guessing” about how animals look like. Initiatives as Biodiversity Heritage Library made accessible huge libraries to many thousands of biologists who, without that, were not able to publish properly. The process of all these technological revolutions is ongoing and improves taxonomy and nomenclature in a way which is unprecedented. All this caused an acceleration in the nomenclatural field: both in quantity and in quality of expertise and fieldwork. The above changes are not without huge problematics. Many studies are carried out on the wide diversity of these problems and even books are written on the subject. -

Organogen.Sis of the Reproductive System of Helisoma Duryi Eudiacus (Pilsbry) (Pulmonata, Gastropoda), with Notes on Breeding Habits
Organogen.sis of the Reproductive System of Helisoma duryi eudiacus (Pilsbry) (Pulmonata, Gastropoda), with Notes on Breeding Habits U~eIM~ Department of Zoology Master of Science ABSTRACT The development of the reproductive system of the frvShwater pulmonate, Helisoma duryi eudiscus (Pilsbry), was studied by means of seriaI sections. Development yas traced from embryos to mature speci mens. Chronological and histologic&l changes in the reproductive organs yere studied. The tyO priaordia which gave ris. to the entire reproductive tract yere clarified. Br.eding habits such as mode of hatching, type of fertilization, quantity of eggs laid, effects of o~gen and temperature on hatching and the resist &Dca of egg membranes to chemicals were additional aspects dealt Yith in this stuay. • Organogenesis of the Reproductive System of He1isoma duryi eudiscus (Pi1sbry) (Pu1monata, Gastropoda), with Notes on Breeding Habits by Mabe1 Mai, B.Sc. A thesis submitted to the Facu1ty of Graduate Studies and Research in partial fu1fi11ment of the requirements for t.he degree of Master of Science. Department of Zoology YcGil1 University, Montreal Ju1y 1969 .' l' r'::\. Mabe1 Mai 19'70 ;1 .i ACKNOWLE1XHŒNTS The wri~er Yi.h•• to thank Dr. Carol M. Lalli for her close supervision over the major part of ~hi. study and for her patience and assistance in rea4ing and correcting every detail of the manuscrip~. A special note of thanks is due to Dr. Tyrell Smith for suggesting the ~opic and supervising part of the experiment. Ur. Ronald Chalk has been Most helptul in providing ~echnical assistance in the preparation of the photomicrographs. -

Kenai National Wildlife Refuge Species List, Version 2018-07-24
Kenai National Wildlife Refuge Species List, version 2018-07-24 Kenai National Wildlife Refuge biology staff July 24, 2018 2 Cover image: map of 16,213 georeferenced occurrence records included in the checklist. Contents Contents 3 Introduction 5 Purpose............................................................ 5 About the list......................................................... 5 Acknowledgments....................................................... 5 Native species 7 Vertebrates .......................................................... 7 Invertebrates ......................................................... 55 Vascular Plants........................................................ 91 Bryophytes ..........................................................164 Other Plants .........................................................171 Chromista...........................................................171 Fungi .............................................................173 Protozoans ..........................................................186 Non-native species 187 Vertebrates ..........................................................187 Invertebrates .........................................................187 Vascular Plants........................................................190 Extirpated species 207 Vertebrates ..........................................................207 Vascular Plants........................................................207 Change log 211 References 213 Index 215 3 Introduction Purpose to avoid implying